The interplay between non-coding RNAs and alternative splicing: from regulatory mechanism to therapeutic implications in cancer
- PMID: 37215575
- PMCID: PMC10196821
- DOI: 10.7150/thno.83920
The interplay between non-coding RNAs and alternative splicing: from regulatory mechanism to therapeutic implications in cancer
Abstract
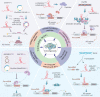
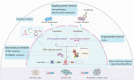
Alternative splicing (AS) is a common and conserved process in eukaryotic gene regulation. It occurs in approximately 95% of multi-exon genes, greatly enriching the complexity and diversity of mRNAs and proteins. Recent studies have found that in addition to coding RNAs, non-coding RNAs (ncRNAs) are also inextricably linked with AS. Multiple different types of ncRNAs are generated by AS of precursor long non-coding (pre-lncRNAs) or precursor messenger RNAs (pre-mRNAs). Furthermore, ncRNAs, as a novel class of regulators, can participate in AS regulation by interacting with the cis-acting elements or trans-acting factors. Several studies have implicated abnormal expression of ncRNAs and ncRNA-related AS events in the initiation, progression, and therapy resistance in various types of cancers. Therefore, owing to their roles in mediating drug resistance, ncRNAs, AS-related factors and AS-related novel antigens may serve as promising therapeutic targets in cancer treatment. In this review, we summarize the interaction between ncRNAs and AS processes, emphasizing their great influences on cancer, especially on chemoresistance, and highlighting their potential values in clinical treatment.
Keywords: Alternative splicing; Cancer; Chemotherapy; Drug resistance; Immunotherapy; Non-coding RNA; Targeted therapy.
© The author(s).
Conflict of interest statement
Competing Interests: The authors have declared that no competing interest exists.
Figures



Similar articles
-
Noncoding RNAs regulate alternative splicing in Cancer.J Exp Clin Cancer Res. 2021 Jan 6;40(1):11. doi: 10.1186/s13046-020-01798-2. J Exp Clin Cancer Res. 2021. PMID: 33407694 Free PMC article. Review.
-
Non-coding RNA in cancer.Essays Biochem. 2021 Oct 27;65(4):625-639. doi: 10.1042/EBC20200032. Essays Biochem. 2021. PMID: 33860799 Free PMC article. Review.
-
Targeting non-coding RNAs to overcome cancer therapy resistance.Signal Transduct Target Ther. 2022 Apr 13;7(1):121. doi: 10.1038/s41392-022-00975-3. Signal Transduct Target Ther. 2022. PMID: 35418578 Free PMC article. Review.
-
ncRNADrug: a database for validated and predicted ncRNAs associated with drug resistance and targeted by drugs.Nucleic Acids Res. 2024 Jan 5;52(D1):D1393-D1399. doi: 10.1093/nar/gkad1042. Nucleic Acids Res. 2024. PMID: 37953323 Free PMC article.
-
Mechanisms of non-coding RNA-modulated alternative splicing in cancer.RNA Biol. 2022;19(1):541-547. doi: 10.1080/15476286.2022.2062846. Epub 2021 Dec 31. RNA Biol. 2022. PMID: 35427215 Free PMC article. Review.
Cited by
-
The Intricate Functional Networks of Pre-mRNA Alternative Splicing in Mammalian Spermatogenesis.Int J Mol Sci. 2024 Nov 10;25(22):12074. doi: 10.3390/ijms252212074. Int J Mol Sci. 2024. PMID: 39596142 Free PMC article. Review.
-
The roles and mechanisms of coding and noncoding RNA variations in cancer.Exp Mol Med. 2024 Sep;56(9):1909-1920. doi: 10.1038/s12276-024-01307-x. Epub 2024 Sep 2. Exp Mol Med. 2024. PMID: 39218979 Free PMC article. Review.
-
The emerging roles of aberrant alternative splicing in glioma.Cell Death Discov. 2025 Feb 6;11(1):50. doi: 10.1038/s41420-025-02323-0. Cell Death Discov. 2025. PMID: 39915450 Free PMC article. Review.
-
New insights into the dynamics of m6A epitranscriptome: hybrid-seq identifies novel mRNAs of the m6A writers METTL3/14.Epigenomics. 2024;16(17):1159-1174. doi: 10.1080/17501911.2024.2390818. Epub 2024 Sep 3. Epigenomics. 2024. PMID: 39225157 Free PMC article.
-
SRSF1-mediated alternative splicing regulates bladder cancer progression and cisplatin sensitivity through HIF1A/BNIP3/mitophagy axis.J Transl Med. 2025 May 22;23(1):571. doi: 10.1186/s12967-025-06547-7. J Transl Med. 2025. PMID: 40405208 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials

