Non-canonical integrin signaling activates EGFR and RAS-MAPK-ERK signaling in small cell lung cancer
- PMID: 37215577
- PMCID: PMC10196829
- DOI: 10.7150/thno.79493
Non-canonical integrin signaling activates EGFR and RAS-MAPK-ERK signaling in small cell lung cancer
Abstract
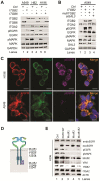
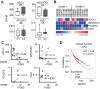
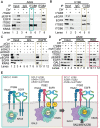
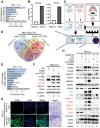
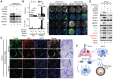
Background: Small cell lung cancer (SCLC) is an extremely aggressive cancer type with a patient median survival of 6-12 months. Epidermal growth factor (EGF) signaling plays an important role in triggering SCLC. In addition, growth factor-dependent signals and alpha-, beta-integrin (ITGA, ITGB) heterodimer receptors functionally cooperate and integrate their signaling pathways. However, the precise role of integrins in EGF receptor (EGFR) activation in SCLC remains elusive. Methods: We analyzed human precision-cut lung slices (hPCLS), retrospectively collected human lung tissue samples and cell lines by classical methods of molecular biology and biochemistry. In addition, we performed RNA-sequencing-based transcriptomic analysis in human lung cancer cells and human lung tissue samples, as well as high-resolution mass spectrometric analysis of the protein cargo from extracellular vesicles (EVs) that were isolated from human lung cancer cells. Results: Our results demonstrate that non-canonical ITGB2 signaling activates EGFR and RAS/MAPK/ERK signaling in SCLC. Further, we identified a novel SCLC gene expression signature consisting of 93 transcripts that were induced by ITGB2, which may be used for stratification of SCLC patients and prognosis prediction of LC patients. We also found a cell-cell communication mechanism based on EVs containing ITGB2, which were secreted by SCLC cells and induced in control human lung tissue RAS/MAPK/ERK signaling and SCLC markers. Conclusions: We uncovered a mechanism of ITGB2-mediated EGFR activation in SCLC that explains EGFR-inhibitor resistance independently of EGFR mutations, suggesting the development of therapies targeting ITGB2 for patients with this extremely aggressive lung cancer type.
Keywords: EGFR; KRAS; extracellular vesicles; integrin; small cell lung cancer.
© The author(s).
Conflict of interest statement
Competing Interests: The authors have declared that no competing interest exists.
Figures



References
-
- Mehta A, Dobersch S, Romero-Olmedo AJ, Barreto G. Epigenetics in lung cancer diagnosis and therapy. Cancer Metastasis Rev. 2015;34(2):229–41. - PubMed
-
- Tartarone A, Giordano P, Lerose R, Rodriquenz MG, Conca R, Aieta M. Progress and challenges in the treatment of small cell lung cancer. Med Oncol. 2017;34:110. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

