The AnnotSV webserver in 2023: updated visualization and ranking
- PMID: 37216590
- PMCID: PMC10320077
- DOI: 10.1093/nar/gkad426
The AnnotSV webserver in 2023: updated visualization and ranking
Abstract
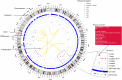
Much of the human genetics variant repertoire is composed of single nucleotide variants (SNV) and small insertion/deletions (indel) but structural variants (SV) remain a major part of our modified DNA. SV detection has often been a complex question to answer either because of the necessity to use different technologies (array CGH, SNP array, Karyotype, Optical Genome Mapping…) to detect each category of SV or to get an appropriate resolution (Whole Genome Sequencing). Thanks to the deluge of pangenomic analysis, Human geneticists are accumulating SV and their interpretation remains time consuming and challenging. The AnnotSV webserver (https://www.lbgi.fr/AnnotSV/) aims at being an efficient tool to (i) annotate and interpret SV potential pathogenicity in the context of human diseases, (ii) recognize potential false positive variants from all the SV identified and (iii) visualize the patient variants repertoire. The most recent developments in the AnnotSV webserver are: (i) updated annotations sources and ranking, (ii) three novel output formats to allow diverse utilization (analysis, pipelines), as well as (iii) two novel user interfaces including an interactive circos view.
© The Author(s) 2023. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures


References
-
- Geoffroy V., Herenger Y., Kress A., Stoetzel C., Piton A., Dollfus H., Muller J.. AnnotSV: an integrated tool for structural variations annotation. Bioinformatics. 2018; 34:3572–3574. - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources

