FOXG1 targets BMP repressors and cell cycle inhibitors in human neural progenitor cells
- PMID: 37216650
- PMCID: PMC10360395
- DOI: 10.1093/hmg/ddad089
FOXG1 targets BMP repressors and cell cycle inhibitors in human neural progenitor cells
Abstract
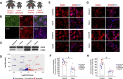
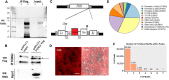
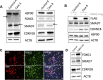
FOXG1 is a critical transcription factor in human brain where loss-of-function mutations cause a severe neurodevelopmental disorder, while increased FOXG1 expression is frequently observed in glioblastoma. FOXG1 is an inhibitor of cell patterning and an activator of cell proliferation in chordate model organisms but different mechanisms have been proposed as to how this occurs. To identify genomic targets of FOXG1 in human neural progenitor cells (NPCs), we engineered a cleavable reporter construct in endogenous FOXG1 and performed chromatin immunoprecipitation (ChIP) sequencing. We also performed deep RNA sequencing of NPCs from two females with loss-of-function mutations in FOXG1 and their healthy biological mothers. Integrative analyses of RNA and ChIP sequencing data showed that cell cycle regulation and Bone Morphogenic Protein (BMP) repression gene ontology categories were over-represented as FOXG1 targets. Using engineered brain cell lines, we show that FOXG1 specifically activates SMAD7 and represses CDKN1B. Activation of SMAD7 which inhibits BMP signaling may be one way that FOXG1 patterns the forebrain, while repression of cell cycle regulators such as CDKN1B may be one way that FOXG1 expands the NPC pool to ensure proper brain size. Our data reveal novel mechanisms on how FOXG1 may control forebrain patterning and cell proliferation in human brain development.
© The Author(s) 2023. Published by Oxford University Press. All rights reserved. For Permissions, please email: journals.permissions@oup.com.
Figures


References
-
- Philippe, C., Amsallem, D., Francannet, C., Lambert, L., Saunier, A., Verneau, F. and Jonveaux, P. (2010) Phenotypic variability in Rett syndrome associated with FOXG1 mutations in females. J. Med. Genet., 47, 59–65. - PubMed
-
- Murphy, D.B., Wiese, S., Burfeind, P., Schmundt, D., Mattei, M.G., Schulz-Schaeffer, W. and Thies, U. (1994) Human brain factor 1, a new member of the fork head gene family. Genomics, 21, 551–557. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

