The genomic landscape of acute lymphoblastic leukemia with intrachromosomal amplification of chromosome 21
- PMID: 37216686
- PMCID: PMC10460677
- DOI: 10.1182/blood.2022019094
The genomic landscape of acute lymphoblastic leukemia with intrachromosomal amplification of chromosome 21
Abstract
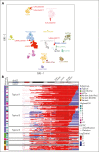
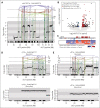
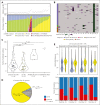
Intrachromosomal amplification of chromosome 21 defines a subtype of high-risk childhood acute lymphoblastic leukemia (iAMP21-ALL) characterized by copy number changes and complex rearrangements of chromosome 21. The genomic basis of iAMP21-ALL and the pathogenic role of the region of amplification of chromosome 21 to leukemogenesis remains incompletely understood. In this study, using integrated whole genome and transcriptome sequencing of 124 patients with iAMP21-ALL, including rare cases arising in the context of constitutional chromosomal aberrations, we identified subgroups of iAMP21-ALL based on the patterns of copy number alteration and structural variation. This large data set enabled formal delineation of a 7.8 Mb common region of amplification harboring 71 genes, 43 of which were differentially expressed compared with non-iAMP21-ALL ones, including multiple genes implicated in the pathogenesis of acute leukemia (CHAF1B, DYRK1A, ERG, HMGN1, and RUNX1). Using multimodal single-cell genomic profiling, including single-cell whole genome sequencing of 2 cases, we documented clonal heterogeneity and genomic evolution, demonstrating that the acquisition of the iAMP21 chromosome is an early event that may undergo progressive amplification during disease ontogeny. We show that UV-mutational signatures and high mutation load are characteristic secondary genetic features. Although the genomic alterations of chromosome 21 are variable, these integrated genomic analyses and demonstration of an extended common minimal region of amplification broaden the definition of iAMP21-ALL for more precise diagnosis using cytogenetic or genomic methods to inform clinical management.
© 2023 by The American Society of Hematology.
Conflict of interest statement
Conflict-of-interest disclosure: C.G.M. received research funding from Loxo Oncology, Pfizer, and AbbVie; honoraria from Amgen and Illumina; and holds stock in Amgen. M.T.R., D.R.B., Z.K., and T.J. are employees of Illumina Inc, a public company that develops and markets systems for genetic analysis. The remaining authors declare no competing financial interests.
Figures




Comment in
-
Insights into the genomics of iAMP21-ALL.Blood. 2023 Aug 24;142(8):682-684. doi: 10.1182/blood.2023021020. Blood. 2023. PMID: 37616022 No abstract available.
References
-
- Moorman AV, Ensor HM, Richards SM, et al. Prognostic effect of chromosomal abnormalities in childhood B-cell precursor acute lymphoblastic leukaemia: results from the UK Medical Research Council ALL97/99 randomised trial. Lancet Oncol. 2010;11(5):429–438. - PubMed
-
- Enshaei A, O'Connor D, Bartram J, et al. A validated novel continuous prognostic index to deliver stratified medicine in pediatric acute lymphoblastic leukemia. Blood. 2020;135(17):1438–1446. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

