Dasatinib overcomes glucocorticoid resistance in B-cell acute lymphoblastic leukemia
- PMID: 37217509
- PMCID: PMC10203345
- DOI: 10.1038/s41467-023-38456-y
Dasatinib overcomes glucocorticoid resistance in B-cell acute lymphoblastic leukemia
Abstract
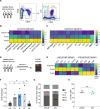
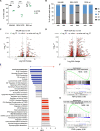
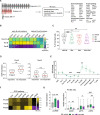
Resistance to glucocorticoids (GC) is associated with an increased risk of relapse in B-cell progenitor acute lymphoblastic leukemia (BCP-ALL). Performing transcriptomic and single-cell proteomic studies in healthy B-cell progenitors, we herein identify coordination between the glucocorticoid receptor pathway with B-cell developmental pathways. Healthy pro-B cells most highly express the glucocorticoid receptor, and this developmental expression is conserved in primary BCP-ALL cells from patients at diagnosis and relapse. In-vitro and in vivo glucocorticoid treatment of primary BCP-ALL cells demonstrate that the interplay between B-cell development and the glucocorticoid pathways is crucial for GC resistance in leukemic cells. Gene set enrichment analysis in BCP-ALL cell lines surviving GC treatment show enrichment of B cell receptor signaling pathways. In addition, primary BCP-ALL cells surviving GC treatment in vitro and in vivo demonstrate a late pre-B cell phenotype with activation of PI3K/mTOR and CREB signaling. Dasatinib, a multi-kinase inhibitor, most effectively targets this active signaling in GC-resistant cells, and when combined with glucocorticoids, results in increased cell death in vitro and decreased leukemic burden and prolonged survival in an in vivo xenograft model. Targeting the active signaling through the addition of dasatinib may represent a therapeutic approach to overcome GC resistance in BCP-ALL.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures



References
Publication types
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

