AMPK is a mechano-metabolic sensor linking cell adhesion and mitochondrial dynamics to Myosin-dependent cell migration
- PMID: 37217519
- PMCID: PMC10202939
- DOI: 10.1038/s41467-023-38292-0
AMPK is a mechano-metabolic sensor linking cell adhesion and mitochondrial dynamics to Myosin-dependent cell migration
Abstract
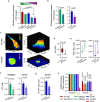
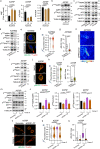
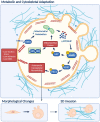
Cell migration is crucial for cancer dissemination. We find that AMP-activated protein kinase (AMPK) controls cell migration by acting as an adhesion sensing molecular hub. In 3-dimensional matrices, fast-migrating amoeboid cancer cells exert low adhesion/low traction linked to low ATP/AMP, leading to AMPK activation. In turn, AMPK plays a dual role controlling mitochondrial dynamics and cytoskeletal remodelling. High AMPK activity in low adhering migratory cells, induces mitochondrial fission, resulting in lower oxidative phosphorylation and lower mitochondrial ATP. Concurrently, AMPK inactivates Myosin Phosphatase, increasing Myosin II-dependent amoeboid migration. Reducing adhesion or mitochondrial fusion or activating AMPK induces efficient rounded-amoeboid migration. AMPK inhibition suppresses metastatic potential of amoeboid cancer cells in vivo, while a mitochondrial/AMPK-driven switch is observed in regions of human tumours where amoeboid cells are disseminating. We unveil how mitochondrial dynamics control cell migration and suggest that AMPK is a mechano-metabolic sensor linking energetics and the cytoskeleton.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures







References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

