Algorithm selection for protein-ligand docking: strategies and analysis on ACE
- PMID: 37217655
- PMCID: PMC10201035
- DOI: 10.1038/s41598-023-35132-5
Algorithm selection for protein-ligand docking: strategies and analysis on ACE
Abstract
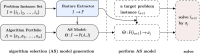
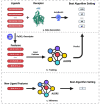
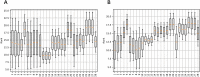
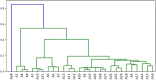
The present study investigates the use of algorithm selection for automatically choosing an algorithm for any given protein-ligand docking task. In drug discovery and design process, conceptualizing protein-ligand binding is a major problem. Targeting this problem through computational methods is beneficial in order to substantially reduce the resource and time requirements for the overall drug development process. One way of addressing protein-ligand docking is to model it as a search and optimization problem. There have been a variety of algorithmic solutions in this respect. However, there is no ultimate algorithm that can efficiently tackle this problem, both in terms of protein-ligand docking quality and speed. This argument motivates devising new algorithms, tailored to the particular protein-ligand docking scenarios. To this end, this paper reports a machine learning-based approach for improved and robust docking performance. The proposed set-up is fully automated, operating without any expert opinion or involvement both on the problem and algorithm aspects. As a case study, an empirical analysis was performed on a well-known protein, Human Angiotensin-Converting Enzyme (ACE), with 1428 ligands. For general applicability, AutoDock 4.2 was used as the docking platform. The candidate algorithms are also taken from AutoDock 4.2. Twenty-eight distinctly configured Lamarckian-Genetic Algorithm (LGA) are chosen to build an algorithm set. ALORS which is a recommender system-based algorithm selection system was preferred for automating the selection from those LGA variants on a per-instance basis. For realizing this selection automation, molecular descriptors and substructure fingerprints were employed as the features characterizing each target protein-ligand docking instance. The computational results revealed that algorithm selection outperforms all those candidate algorithms. Further assessment is reported on the algorithms space, discussing the contributions of LGA's parameters. As it pertains to protein-ligand docking, the contributions of the aforementioned features are examined, which shed light on the critical features affecting the docking performance.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures





References
-
- Everhardus JA. Drug Design: Medicinal Chemistry. Elsevier; 2017.
-
- Edgar L-L, Jurgen B, Jose LM-F. Informatics for chemistry, biology, and biomedical sciences. J. Chem. Inf. Model. 2020;61(1):26–35. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

