Immunovirological and environmental screening reveals actionable risk factors for fatal COVID-19 during post-vaccination nursing home outbreaks
- PMID: 37217661
- PMCID: PMC10275758
- DOI: 10.1038/s43587-023-00421-1
Immunovirological and environmental screening reveals actionable risk factors for fatal COVID-19 during post-vaccination nursing home outbreaks
Abstract
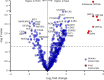
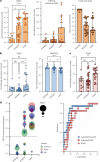
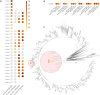
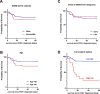
Coronavirus Disease 2019 (COVID-19) vaccination has resulted in excellent protection against fatal disease, including in older adults. However, risk factors for post-vaccination fatal COVID-19 are largely unknown. We comprehensively studied three large nursing home outbreaks (20-35% fatal cases among residents) by combining severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) aerosol monitoring, whole-genome phylogenetic analysis and immunovirological profiling of nasal mucosa by digital nCounter transcriptomics. Phylogenetic investigations indicated that each outbreak stemmed from a single introduction event, although with different variants (Delta, Gamma and Mu). SARS-CoV-2 was detected in aerosol samples up to 52 d after the initial infection. Combining demographic, immune and viral parameters, the best predictive models for mortality comprised IFNB1 or age, viral ORF7a and ACE2 receptor transcripts. Comparison with published pre-vaccine fatal COVID-19 transcriptomic and genomic signatures uncovered a unique IRF3 low/IRF7 high immune signature in post-vaccine fatal COVID-19 outbreaks. A multi-layered strategy, including environmental sampling, immunomonitoring and early antiviral therapy, should be considered to prevent post-vaccination COVID-19 mortality in nursing homes.
© 2023. The Author(s).
Conflict of interest statement
K.L. received consultancy fees from MRM Health and Merck Sharp & Dohme, speaker fees from Pfizer and Gilead and service fees from Thermo Fisher Scientific and TECOmedical, all outside the reported work. The other authors report no competing interests.
Figures




References
-
- Hardy JO, et al. A world apart: levels and determinants of excess mortality due to COVID-19 in care homes: the case of the Belgian region of Wallonia during the spring 2020 wave. Demogr. Res. 2021;45:1011–1040. doi: 10.4054/DemRes.2021.45.33. - DOI
-
- Gillain S, Belche J-L, Moreau J-F. COVID-19 epidemic in the nursing homes in Belgium. J. Nursing Home Res. 2020;6:40–42.
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

