Spatial imaging of glycoRNA in single cells with ARPLA
- PMID: 37217750
- PMCID: PMC10663388
- DOI: 10.1038/s41587-023-01801-z
Spatial imaging of glycoRNA in single cells with ARPLA
Abstract
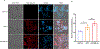
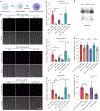
Little is known about the biological roles of glycosylated RNAs (glycoRNAs), a recently discovered class of glycosylated molecules, because of a lack of visualization methods. We report sialic acid aptamer and RNA in situ hybridization-mediated proximity ligation assay (ARPLA) to visualize glycoRNAs in single cells with high sensitivity and selectivity. The signal output of ARPLA occurs only when dual recognition of a glycan and an RNA triggers in situ ligation, followed by rolling circle amplification of a complementary DNA, which generates a fluorescent signal by binding fluorophore-labeled oligonucleotides. Using ARPLA, we detect spatial distributions of glycoRNAs on the cell surface and their colocalization with lipid rafts as well as the intracellular trafficking of glycoRNAs through SNARE protein-mediated secretory exocytosis. Studies in breast cell lines suggest that surface glycoRNA is inversely associated with tumor malignancy and metastasis. Investigation of the relationship between glycoRNAs and monocyte-endothelial cell interactions suggests that glycoRNAs may mediate cell-cell interactions during the immune response.
© 2023. The Author(s), under exclusive licence to Springer Nature America, Inc.
Figures









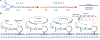



Comment in
-
ARPLA for spatial imaging of glycoRNAs.Nat Rev Cancer. 2023 Oct;23(10):651. doi: 10.1038/s41568-023-00614-1. Nat Rev Cancer. 2023. PMID: 37644184 No abstract available.
References
-
- Caldwell RM & Flynn RA Discovering glycoRNA: traditional and non-canonical approaches to studying RNA modifications. Isr. J. Chem 63, e202200059 (2022).
-
- Suzuki T. The expanding world of tRNA modifications and their disease relevance. Nat. Rev. Mol. Cell Biol 22, 375–392 (2021). - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

