In vitro selection of macrocyclic peptide inhibitors containing cyclic γ2,4-amino acids targeting the SARS-CoV-2 main protease
- PMID: 37217786
- PMCID: PMC10322702
- DOI: 10.1038/s41557-023-01205-1
In vitro selection of macrocyclic peptide inhibitors containing cyclic γ2,4-amino acids targeting the SARS-CoV-2 main protease
Abstract
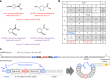
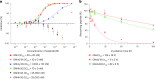
γ-Amino acids can play important roles in the biological activities of natural products; however, the ribosomal incorporation of γ-amino acids into peptides is challenging. Here we report how a selection campaign employing a non-canonical peptide library containing cyclic γ2,4-amino acids resulted in the discovery of very potent inhibitors of the SARS-CoV-2 main protease (Mpro). Two kinds of cyclic γ2,4-amino acids, cis-3-aminocyclobutane carboxylic acid (γ1) and (1R,3S)-3-aminocyclopentane carboxylic acid (γ2), were ribosomally introduced into a library of thioether-macrocyclic peptides. One resultant potent Mpro inhibitor (half-maximal inhibitory concentration = 50 nM), GM4, comprising 13 residues with γ1 at the fourth position, manifests a 5.2 nM dissociation constant. An Mpro:GM4 complex crystal structure reveals the intact inhibitor spans the substrate binding cleft. The γ1 interacts with the S1' catalytic subsite and contributes to a 12-fold increase in proteolytic stability compared to its alanine-substituted variant. Knowledge of interactions between GM4 and Mpro enabled production of a variant with a 5-fold increase in potency.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures




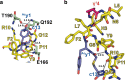

References
-
- Hintermann T, Gademann K, Jaun B, Seebach D. γ-Peptides forming more stable secondary structures than α-peptides: synthesis and helical NMR-solution structure of the γ-hexapeptide analog of H-(Val-Ala-Leu)2-OH. Helv. Chim. Acta. 1998;81:983–1002. doi: 10.1002/hlca.19980810514. - DOI
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

