The role of plasmids in carbapenem resistant E. coli in Alameda County, California
- PMID: 37217873
- PMCID: PMC10201492
- DOI: 10.1186/s12866-023-02900-2
The role of plasmids in carbapenem resistant E. coli in Alameda County, California
Abstract
Background: Antimicrobial resistant infections continue to be a leading global public health crisis. Mobile genetic elements, such as plasmids, have been shown to play a major role in the dissemination of antimicrobial resistance (AMR) genes. Despite its ongoing threat to human health, surveillance of AMR in the United States is often limited to phenotypic resistance. Genomic analyses are important to better understand the underlying resistance mechanisms, assess risk, and implement appropriate prevention strategies. This study aimed to investigate the extent of plasmid mediated antimicrobial resistance that can be inferred from short read sequences of carbapenem resistant E. coli (CR-Ec) in Alameda County, California. E. coli isolates from healthcare locations in Alameda County were sequenced using an Illumina MiSeq and assembled with Unicycler. Genomes were categorized according to predefined multilocus sequence typing (MLST) and core genome multilocus sequence typing (cgMLST) schemes. Resistance genes were identified and corresponding contigs were predicted to be plasmid-borne or chromosome-borne using two bioinformatic tools (MOB-suite and mlplasmids).
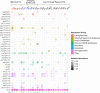
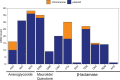
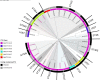
Results: Among 82 of CR-Ec identified between 2017 and 2019, twenty-five sequence types (STs) were detected. ST131 was the most prominent (n = 17) followed closely by ST405 (n = 12). blaCTX-M were the most common ESBL genes and just over half (18/30) of these genes were predicted to be plasmid-borne by both MOB-suite and mlplasmids. Three genetically related groups of E. coli isolates were identified with cgMLST. One of the groups contained an isolate with a chromosome-borne blaCTX-M-15 gene and an isolate with a plasmid-borne blaCTX-M-15 gene.
Conclusions: This study provides insights into the dominant clonal groups driving carbapenem resistant E. coli infections in Alameda County, CA, USA clinical sites and highlights the relevance of whole-genome sequencing in routine local genomic surveillance. The finding of multi-drug resistant plasmids harboring high-risk resistance genes is of concern as it indicates a risk of dissemination to previously susceptible clonal groups, potentially complicating clinical and public health intervention.
Keywords: Carbapenem resistance; ESBLs; Plasmids; Short-read whole genome sequencing; bla CTX−M−15.
© 2023. The Author(s).
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
References
-
- Pulingam T, Parumasivam T, Gazzali AM, Sulaiman AM, Chee JY, Lakshmanan M, et al. Antimicrobial resistance: prevalence, economic burden, mechanisms of resistance and strategies to overcome. Eur J Pharm Sci. 2022 Mar;170:106103. - PubMed
-
- Centers for Disease Control and Prevention (U.S.). Antibiotic resistance threats in the United States, 2019 [Internet]. Centers for Disease Control and Prevention (U.S.); 2019 Nov [cited 2022 Dec 30]. Available from: https://stacks.cdc.gov/view/cdc/82532.

