The BET PROTAC inhibitor dBET6 protects against retinal degeneration and inhibits the cGAS-STING in response to light damage
- PMID: 37217935
- PMCID: PMC10201800
- DOI: 10.1186/s12974-023-02804-y
The BET PROTAC inhibitor dBET6 protects against retinal degeneration and inhibits the cGAS-STING in response to light damage
Abstract
Background: Chronic inflammation significantly contributes to photoreceptor death in blinding retinal diseases such as age-related macular degeneration (AMD) and retinitis pigmentosa (RP). Bromodomain and extraterminal domain (BET) proteins are epigenetic readers that act as key proinflammatory factors. We recently found the first-generation BET inhibitor JQ1 alleviated sodium iodate-induced retinal degeneration by suppressing cGAS-STING innate immunity. Here, we investigated the effects and mechanism of dBET6, a proteolysis‑targeting chimera (PROTAC) small molecule that selectively degrades BET by the ubiquitin‒proteasome system, in light-induced retinal degeneration.
Methods: Mice were exposed to bright light to induce retinal degeneration, and the activation of cGAS-STING was determined by RNA-sequencing and molecular biology. Retinal function, morphology, photoreceptor viability and retinal inflammation were examined in the presence and absence of dBET6 treatment.
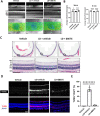
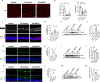
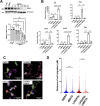
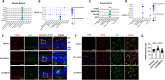
Results: Intraperitoneal injection of dBET6 led to the rapid degradation of BET protein in the retina without detectable toxicity. dBET6 improved retinal responsiveness and visual acuity after light damage (LD). dBET6 also repressed LD-induced retinal macrophages/microglia activation, Müller cell gliosis, photoreceptor death and retinal degeneration. Analysis of single-cell RNA-sequencing results revealed cGAS-STING components were expressed in retinal microglia. LD led to dramatic activation of the cGAS-STING pathway, whereas dBET6 suppressed LD-induced STING expression in reactive macrophages/microglia and the related inflammatory response.
Conclusions: This study indicates targeted degradation of BET by dBET6 exerts neuroprotective effects by inhibiting cGAS-STING in reactive retinal macrophages/microglia, and is expected to become a new strategy for treatment of retinal degeneration.
Keywords: BET; Microglia/macrophage; Neuroinflammation; PROTAC; Retinal degeneration; cGAS-STING.
© 2023. The Author(s).
Conflict of interest statement
The authors declare that they have no competing interests.
Figures





References
-
- Pfau M, von der Emde L, de Sisternes L, Hallak JA, Leng T, Schmitz-Valckenberg S, Holz FG, Fleckenstein M, Rubin DL. Progression of photoreceptor degeneration in geographic atrophy secondary to age-related macular degeneration. JAMA Ophthalmol. 2020;138:1026–1034. doi: 10.1001/jamaophthalmol.2020.2914. - DOI - PMC - PubMed
-
- Hume DA, Perry VH, Gordon S. Immunohistochemical localization of a macrophage-specific antigen in developing mouse retina: phagocytosis of dying neurons and differentiation of microglial cells to form a regular array in the plexiform layers. J Cell Biol. 1983;97:253–257. doi: 10.1083/jcb.97.1.253. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

