Functional and evolutionary study of MLO gene family in the regulation of Sclerotinia stem rot resistance in Brassica napus L
- PMID: 37217949
- PMCID: PMC10204302
- DOI: 10.1186/s13068-023-02325-z
Functional and evolutionary study of MLO gene family in the regulation of Sclerotinia stem rot resistance in Brassica napus L
Abstract
Background: Oilseed rape (Brassica napus L.) is known as one of the most important oilseed crops cultivated around the world. However, its production continuously faces a huge challenge of Sclerotinia stem rot (SSR), a destructive disease caused by the fungus Sclerotinia sclerotiorum, resulting in huge yield loss annually. The SSR resistance in B. napus is quantitative and controlled by a set of minor genes. Identification of these genes and pyramiding them into a variety are a major strategy for SSR resistance breeding in B. napus.
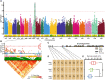
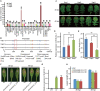
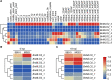
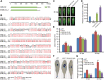
Results: Here, we performed a genome-wide association study (GWAS) using a natural population of B. napus consisting of 222 accessions to identify BnaA08g25340D (BnMLO2_2) as a candidate gene that regulates the SSR resistance. BnMLO2_2 was a member of seven homolog genes of Arabidopsis Mildew Locus O 2 (MLO2) and the significantly SNPs were mainly distributed in the promoter of BnMLO2_2, suggesting a role of BnMLO2_2 expression level in the regulation of SSR resistance. We expressed BnMLO2_2 in Arabidopsis and the transgenic plants displayed an enhanced SSR resistance. Transcriptome profiling of different tissues of B. napus revealed that BnMLO2_2 had the most expression level in leaf and silique tissues among all the 7 BnMLO2 members and also expressed higher in the SSR resistant accession than in the susceptible accession. In Arabidopsis, mlo2 plants displayed reduced resistance to SSR, whereas overexpression of MLO2 conferred plants an enhanced SSR resistance. Moreover, a higher expression level of MLO2 showed a stronger SSR resistance in the transgenic plants. The regulation of MLO2 in SSR resistance may be associated with the cell death. Collinearity and phylogenetic analysis revealed a large expansion of MLO family in Brassica crops.
Conclusion: Our study revealed an important role of BnMLO2 in the regulation of SSR resistance and provided a new gene candidate for future improvement of SSR resistance in B. napus and also new insights into understanding of MLO family evolution in Brassica crops.
Keywords: Brassica napus L.; Evolution; Gene expression; Genome-wide association studies; MLO; Sclerotinia stem rot; Transcriptome.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures
Similar articles
-
Genome-wide Association Study Identifies New Loci for Resistance to Sclerotinia Stem Rot in Brassica napus.Front Plant Sci. 2016 Sep 20;7:1418. doi: 10.3389/fpls.2016.01418. eCollection 2016. Front Plant Sci. 2016. PMID: 27703464 Free PMC article.
-
Screening of microRNAs and target genes involved in Sclerotinia sclerotiorum (Lib.) infection in Brassica napus L.BMC Plant Biol. 2023 Oct 9;23(1):479. doi: 10.1186/s12870-023-04501-7. BMC Plant Biol. 2023. PMID: 37807039 Free PMC article.
-
Arabidopsis GDSL1 overexpression enhances rapeseed Sclerotinia sclerotiorum resistance and the functional identification of its homolog in Brassica napus.Plant Biotechnol J. 2020 May;18(5):1255-1270. doi: 10.1111/pbi.13289. Epub 2019 Nov 20. Plant Biotechnol J. 2020. PMID: 31693306 Free PMC article.
-
Sclerotinia Stem Rot Resistance in Rapeseed: Recent Progress and Future Prospects.J Agric Food Chem. 2021 Mar 17;69(10):2965-2978. doi: 10.1021/acs.jafc.0c07351. Epub 2021 Mar 5. J Agric Food Chem. 2021. PMID: 33667087 Review.
-
Current Status and Challenges in Identifying Disease Resistance Genes in Brassica napus.Front Plant Sci. 2017 Nov 6;8:1788. doi: 10.3389/fpls.2017.01788. eCollection 2017. Front Plant Sci. 2017. PMID: 29163558 Free PMC article. Review.
Cited by
-
Unraveling the Intricacies of Powdery Mildew: Insights into Colonization, Plant Defense Mechanisms, and Future Strategies.Int J Mol Sci. 2025 Apr 9;26(8):3513. doi: 10.3390/ijms26083513. Int J Mol Sci. 2025. PMID: 40331988 Free PMC article. Review.
-
Major Genes for Powdery Mildew Resistance in Research and Breeding of Barley: A Few Brief Narratives and Recommendations.Plants (Basel). 2025 Jul 8;14(14):2091. doi: 10.3390/plants14142091. Plants (Basel). 2025. PMID: 40733328 Free PMC article. Review.
-
Integrated Assays of Genome-Wide Association Study, Multi-Omics Co-Localization, and Machine Learning Associated Calcium Signaling Genes with Oilseed Rape Resistance to Sclerotinia sclerotiorum.Int J Mol Sci. 2024 Jun 25;25(13):6932. doi: 10.3390/ijms25136932. Int J Mol Sci. 2024. PMID: 39000053 Free PMC article.
References
-
- Friedt W, Tu J, Fu T. Academic and economic importance of brassica napus rapeseed. In: Liu S, Snowdon R, Chalhoub B, editors. The Brassica napus Genome. Cham: Springer International Publishing; 2018. pp. 1–20.
-
- Felten D, Fröba N, Fries J, Emmerling C. Energy balances and greenhouse gas-mitigation potentials of bioenergy cropping systems (Miscanthus, rapeseed, and maize) based on farming conditions in Western Germany. Renew Energy. 2013;55:160–174. doi: 10.1016/j.renene.2012.12.004. - DOI
-
- D'Avino L, Dainelli R, Lazzeri L, Spugnoli P. The role of co-products in biorefinery sustainability: energy allocation versus substitution method in rapeseed and carinata biodiesel chains. J Clean Prod. 2015;94:108. doi: 10.1016/j.jclepro.2015.01.088. - DOI
-
- Morinaga T. Interspecific hybridization in brassica:VI. the cytology of F1 hybrids of B. juncea and B. nigra. Cytologia. 1934;6:62–67. doi: 10.1508/cytologia.6.62. - DOI
-
- Purdy LH. Sclerotinia sclerotiorum: history, diseases and symptomatology, host range, geographic distribution, and impact. Phytopathology. 1979;69:875–880. doi: 10.1094/Phyto-69-875. - DOI
Grants and funding
LinkOut - more resources
Full Text Sources
