Coupled protein quality control during nonsense-mediated mRNA decay
- PMID: 37218462
- PMCID: PMC10234110
- DOI: 10.1242/jcs.261216
Coupled protein quality control during nonsense-mediated mRNA decay
Abstract
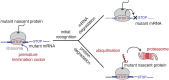
Translation of mRNAs containing premature termination codons (PTCs) results in truncated protein products with deleterious effects. Nonsense-mediated decay (NMD) is a surveillance pathway responsible for detecting PTC containing transcripts. Although the molecular mechanisms governing mRNA degradation have been extensively studied, the fate of the nascent protein product remains largely uncharacterized. Here, we use a fluorescent reporter system in mammalian cells to reveal a selective degradation pathway specifically targeting the protein product of an NMD mRNA. We show that this process is post-translational and dependent on the ubiquitin proteasome system. To systematically uncover factors involved in NMD-linked protein quality control, we conducted genome-wide flow cytometry-based screens. Our screens recovered known NMD factors but suggested that protein degradation did not depend on the canonical ribosome-quality control (RQC) pathway. A subsequent arrayed screen demonstrated that protein and mRNA branches of NMD rely on a shared recognition event. Our results establish the existence of a targeted pathway for nascent protein degradation from PTC containing mRNAs, and provide a reference for the field to identify and characterize required factors.
Keywords: Nonsense-mediated decay; Quality control; Ubiquitin-proteasome pathway; mRNA.
© 2023. Published by The Company of Biologists Ltd.
Conflict of interest statement
Competing interests J.S.W. declares outside interest in 5 AM Venture, Amgen, Chroma Medicine, KSQ Therapeutics, Maze Therapeutics, Tenaya Therapeutics, Tessera Therapeutics, and Third Rock Ventures. R.M.V. declares outside interest in Gate Biosciences.
Figures


Similar articles
-
Selective destabilization of polypeptides synthesized from NMD-targeted transcripts.Mol Biol Cell. 2021 Dec 1;32(22):ar38. doi: 10.1091/mbc.E21-08-0382. Epub 2021 Sep 29. Mol Biol Cell. 2021. PMID: 34586879 Free PMC article.
-
Cellular variability of nonsense-mediated mRNA decay.Nat Commun. 2021 Dec 10;12(1):7203. doi: 10.1038/s41467-021-27423-0. Nat Commun. 2021. PMID: 34893608 Free PMC article.
-
Arginine CGA codons as a source of nonsense mutations: a possible role in multivariant gene expression, control of mRNA quality, and aging.Mol Genet Genomics. 2017 Oct;292(5):1013-1026. doi: 10.1007/s00438-017-1328-y. Epub 2017 May 18. Mol Genet Genomics. 2017. PMID: 28523359
-
Nonsense-mediated mRNA decay: an intricate machinery that shapes transcriptomes.Nat Rev Mol Cell Biol. 2015 Nov;16(11):665-77. doi: 10.1038/nrm4063. Epub 2015 Sep 23. Nat Rev Mol Cell Biol. 2015. PMID: 26397022 Review.
-
The broader sense of nonsense.Trends Biochem Sci. 2022 Nov;47(11):921-935. doi: 10.1016/j.tibs.2022.06.003. Epub 2022 Jun 29. Trends Biochem Sci. 2022. PMID: 35780009 Review.
Cited by
-
Mechanisms and Delivery of tRNA Therapeutics.Chem Rev. 2024 Jun 26;124(12):7976-8008. doi: 10.1021/acs.chemrev.4c00142. Epub 2024 May 27. Chem Rev. 2024. PMID: 38801719 Free PMC article. Review.
-
Mutations causing premature termination codons discriminate and generate cellular and clinical variability in HHT.Blood. 2024 May 30;143(22):2314-2331. doi: 10.1182/blood.2023021777. Blood. 2024. PMID: 38457357 Free PMC article.
-
A system of reporters for comparative investigation of EJC-independent and EJC-enhanced nonsense-mediated mRNA decay.Nucleic Acids Res. 2024 Apr 12;52(6):e34. doi: 10.1093/nar/gkae121. Nucleic Acids Res. 2024. PMID: 38375914 Free PMC article.
-
Nonsense-Mediated mRNA Decay Factor Functions in Human Health and Disease.Biomedicines. 2023 Feb 27;11(3):722. doi: 10.3390/biomedicines11030722. Biomedicines. 2023. PMID: 36979701 Free PMC article. Review.
-
Ending a bad start: Triggers and mechanisms of co-translational protein degradation.Front Mol Biosci. 2023 Jan 4;9:1089825. doi: 10.3389/fmolb.2022.1089825. eCollection 2022. Front Mol Biosci. 2023. PMID: 36660423 Free PMC article. Review.
References
-
- Andersen, C. B. F., Ballut, L., Johansen, J. S., Chamieh, H., Nielsen, K. H., Olliveira, C. L. P., Pedersen, J. S., Seraphin, B., Le Hir, H. E. and Andersen, G. R. (2006). Structure of the exon junction core complex with a trapped DEAD-box ATPase bound RNA. Science 313, 1968-1972. 10.1126/science.1131981 - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

