Tumor-activated lymph node fibroblasts suppress T cell function in diffuse large B cell lymphoma
- PMID: 37219943
- PMCID: PMC10313378
- DOI: 10.1172/JCI166070
Tumor-activated lymph node fibroblasts suppress T cell function in diffuse large B cell lymphoma
Abstract
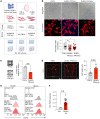
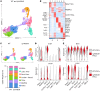
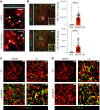
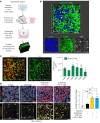
Recent transcriptomic-based analysis of diffuse large B cell lymphoma (DLBCL) has highlighted the clinical relevance of LN fibroblast and tumor-infiltrating lymphocyte (TIL) signatures within the tumor microenvironment (TME). However, the immunomodulatory role of fibroblasts in lymphoma remains unclear. Here, by studying human and mouse DLBCL-LNs, we identified the presence of an aberrantly remodeled fibroblastic reticular cell (FRC) network expressing elevated fibroblast-activated protein (FAP). RNA-Seq analyses revealed that exposure to DLBCL reprogrammed key immunoregulatory pathways in FRCs, including a switch from homeostatic to inflammatory chemokine expression and elevated antigen-presentation molecules. Functional assays showed that DLBCL-activated FRCs (DLBCL-FRCs) hindered optimal TIL and chimeric antigen receptor (CAR) T cell migration. Moreover, DLBCL-FRCs inhibited CD8+ TIL cytotoxicity in an antigen-specific manner. Notably, the interrogation of patient LNs with imaging mass cytometry identified distinct environments differing in their CD8+ TIL-FRC composition and spatial organization that associated with survival outcomes. We further demonstrated the potential to target inhibitory FRCs to rejuvenate interacting TILs. Cotreating organotypic cultures with FAP-targeted immunostimulatory drugs and a bispecific antibody (glofitamab) augmented antilymphoma TIL cytotoxicity. Our study reveals an immunosuppressive role of FRCs in DLBCL, with implications for immune evasion, disease pathogenesis, and optimizing immunotherapy for patients.
Keywords: Cancer immunotherapy; Immunology; Lymphomas; Oncology.
Conflict of interest statement
Figures









Comment in
-
Discovering the fibroblastic reticular cell in the immune tumor microenvironment in lymphoma.J Clin Invest. 2023 Jul 3;133(13):e171310. doi: 10.1172/JCI171310. J Clin Invest. 2023. PMID: 37395274 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

