Omicron BQ.1.1 and XBB.1 unprecedentedly escape broadly neutralizing antibodies elicited by prototype vaccination
- PMID: 37219999
- PMCID: PMC10201307
- DOI: 10.1016/j.celrep.2023.112532
Omicron BQ.1.1 and XBB.1 unprecedentedly escape broadly neutralizing antibodies elicited by prototype vaccination
Abstract
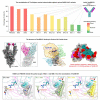
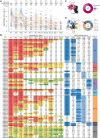
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) Omicron subvariants have seriously attacked the antibody barrier established by natural infection and/or vaccination, especially the recently emerged BQ.1.1 and XBB.1. However, crucial mechanisms underlying the virus escape and the broad neutralization remain elusive. Here, we present a panoramic analysis of broadly neutralizing activity and binding epitopes of 75 monoclonal antibodies isolated from prototype inactivated vaccinees. Nearly all neutralizing antibodies (nAbs) partly or totally lose their neutralization against BQ.1.1 and XBB.1. We report a broad nAb, VacBB-551, that effectively neutralizes all tested subvariants including BA.2.75, BQ.1.1, and XBB.1. We determine the cryoelectron microscopy (cryo-EM) structure of VacBB-551 complexed with the BA.2 spike and perform detailed functional verification to reveal the molecular basis of N460K and F486V/S mutations mediating the partial escape of BA.2.75, BQ.1.1, and XBB.1 from the neutralization of VacBB-551. Overall, BQ.1.1 and XBB.1 raised the alarm over SARS-CoV-2 evolution with unprecedented antibody evasion from broad nAbs elicited by prototype vaccination.
Keywords: BQ.1.1; CP: Immunology; CP: Microbiology; Omicron subvariant; SARS-CoV-2; XBB.1; broadly neutralizing activity; structure analysis.
Copyright © 2023 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests Patent applications have been filed on some of mAbs presented here. Z.Z., B.J., Q.F., C.L., S.S., M.W., B.Z., and X.G. are inventors.
Figures




References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

