gExcite: a start-to-end framework for single-cell gene expression, hashing, and antibody analysis
- PMID: 37220897
- PMCID: PMC10229235
- DOI: 10.1093/bioinformatics/btad329
gExcite: a start-to-end framework for single-cell gene expression, hashing, and antibody analysis
Abstract
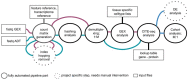
Summary: Recently, CITE-seq emerged as a multimodal single-cell technology capturing gene expression and surface protein information from the same single cells, which allows unprecedented insights into disease mechanisms and heterogeneity, as well as immune cell profiling. Multiple single-cell profiling methods exist, but they are typically focused on either gene expression or antibody analysis, not their combination. Moreover, existing software suites are not easily scalable to a multitude of samples. To this end, we designed gExcite, a start-to-end workflow that provides both gene and antibody expression analysis, as well as hashing deconvolution. Embedded in the Snakemake workflow manager, gExcite facilitates reproducible and scalable analyses. We showcase the output of gExcite on a study of different dissociation protocols on PBMC samples.
Availability and implementation: gExcite is open source available on github at https://github.com/ETH-NEXUS/gExcite_pipeline. The software is distributed under the GNU General Public License 3 (GPL3).
© The Author(s) 2023. Published by Oxford University Press.
Figures
References
-
- Kim HJ, Lin Y, Geddes TA. et al. CiteFuse enables multi-modal analysis of CITE-Seq data. Bioinformatics 2020;36:4137–43. - PubMed