An mRNA-based T-cell-inducing antigen strengthens COVID-19 vaccine against SARS-CoV-2 variants
- PMID: 37221158
- PMCID: PMC10204679
- DOI: 10.1038/s41467-023-38751-8
An mRNA-based T-cell-inducing antigen strengthens COVID-19 vaccine against SARS-CoV-2 variants
Abstract
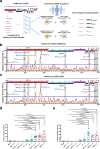
Herd immunity achieved through mass vaccination is an effective approach to prevent contagious diseases. Nonetheless, emerging SARS-CoV-2 variants with frequent mutations largely evaded humoral immunity induced by Spike-based COVID-19 vaccines. Herein, we develop a lipid nanoparticle (LNP)-formulated mRNA-based T-cell-inducing antigen, which targeted three SARS-CoV-2 proteome regions that enriched human HLA-I epitopes (HLA-EPs). Immunization of HLA-EPs induces potent cellular responses to prevent SARS-CoV-2 infection in humanized HLA-A*02:01/DR1 and HLA-A*11:01/DR1 transgenic mice. Of note, the sequences of HLA-EPs are highly conserved among SARS-CoV-2 variants of concern. In humanized HLA-transgenic mice and female rhesus macaques, dual immunization with the LNP-formulated mRNAs encoding HLA-EPs and the receptor-binding domain of the SARS-CoV-2 B.1.351 variant (RBDbeta) is more efficacious in preventing infection of SARS-CoV-2 Beta and Omicron BA.1 variants than single immunization of LNP-RBDbeta. This study demonstrates the necessity to strengthen the vaccine effectiveness by comprehensively stimulating both humoral and cellular responses, thereby offering insight for optimizing the design of COVID-19 vaccines.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures



References
-
- World Health Organization. Weekly epidemiological update on COVID-19. https://www.who.int/publications/m/item/weekly-epidemiological-update-on... (2023).
Publication types
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

