Targeting small GTPases: emerging grasps on previously untamable targets, pioneered by KRAS
- PMID: 37221195
- PMCID: PMC10205766
- DOI: 10.1038/s41392-023-01441-4
Targeting small GTPases: emerging grasps on previously untamable targets, pioneered by KRAS
Abstract
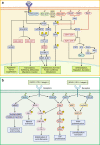
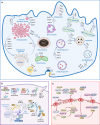
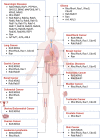
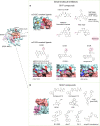
Small GTPases including Ras, Rho, Rab, Arf, and Ran are omnipresent molecular switches in regulating key cellular functions. Their dysregulation is a therapeutic target for tumors, neurodegeneration, cardiomyopathies, and infection. However, small GTPases have been historically recognized as "undruggable". Targeting KRAS, one of the most frequently mutated oncogenes, has only come into reality in the last decade due to the development of breakthrough strategies such as fragment-based screening, covalent ligands, macromolecule inhibitors, and PROTACs. Two KRASG12C covalent inhibitors have obtained accelerated approval for treating KRASG12C mutant lung cancer, and allele-specific hotspot mutations on G12D/S/R have been demonstrated as viable targets. New methods of targeting KRAS are quickly evolving, including transcription, immunogenic neoepitopes, and combinatory targeting with immunotherapy. Nevertheless, the vast majority of small GTPases and hotspot mutations remain elusive, and clinical resistance to G12C inhibitors poses new challenges. In this article, we summarize diversified biological functions, shared structural properties, and complex regulatory mechanisms of small GTPases and their relationships with human diseases. Furthermore, we review the status of drug discovery for targeting small GTPases and the most recent strategic progress focused on targeting KRAS. The discovery of new regulatory mechanisms and development of targeting approaches will together promote drug discovery for small GTPases.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures



References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

