Using system biology and bioinformatics to identify the influences of COVID-19 co-infection with influenza virus on COPD
- PMID: 37221323
- PMCID: PMC10205564
- DOI: 10.1007/s10142-023-01091-3
Using system biology and bioinformatics to identify the influences of COVID-19 co-infection with influenza virus on COPD
Abstract
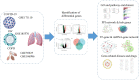
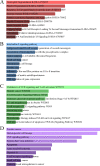
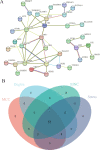
Coronavirus disease 2019 (COVID-19) has speedily increased mortality globally. Although they are risk factors for severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), less is known about the common molecular mechanisms behind COVID-19, influenza virus A (IAV), and chronic obstructive pulmonary disease (COPD). This research used bioinformatics and systems biology to find possible medications for treating COVID-19, IAV, and COPD via identifying differentially expressed genes (DEGs) from gene expression datasets (GSE171110, GSE76925, GSE106986, and GSE185576). A total of 78 DEGs were subjected to functional enrichment, pathway analysis, protein-protein interaction (PPI) network construct, hub gene extraction, and other potentially relevant disorders. Then, DEGs were discovered in networks including transcription factor (TF)-gene connections, protein-drug interactions, and DEG-microRNA (miRNA) coregulatory networks by using NetworkAnalyst. The top 12 hub genes were MPO, MMP9, CD8A, HP, ELANE, CD5, CR2, PLA2G7, PIK3R1, SLAMF1, PEX3, and TNFRSF17. We found that 44 TFs-genes, as well as 118 miRNAs, are directly linked to hub genes. Additionally, we searched the Drug Signatures Database (DSigDB) and identified 10 drugs that could potentially treat COVID-19, IAV, and COPD. Therefore, we evaluated the top 12 hub genes that could be promising DEGs for targeted therapy for SARS-CoV-2 and identified several prospective medications that may benefit COPD patients with COVID-19 and IAV co-infection.
Keywords: COPD; COVID-19; Differentially expressed genes; Drug molecule; Hub genes; Influenza viruses.
© 2023. The Author(s), under exclusive licence to Springer-Verlag GmbH Germany, part of Springer Nature.
Conflict of interest statement
The authors declare no competing interests.
Figures



Similar articles
-
Network-Based Data Analysis Reveals Ion Channel-Related Gene Features in COVID-19: A Bioinformatic Approach.Biochem Genet. 2023 Apr;61(2):471-505. doi: 10.1007/s10528-022-10280-x. Epub 2022 Sep 14. Biochem Genet. 2023. PMID: 36104591 Free PMC article.
-
Bioinformatics and system biology approach to identify the influences of SARS-CoV-2 infections to idiopathic pulmonary fibrosis and chronic obstructive pulmonary disease patients.Brief Bioinform. 2021 Sep 2;22(5):bbab115. doi: 10.1093/bib/bbab115. Brief Bioinform. 2021. PMID: 33847347 Free PMC article.
-
Bioinformatics and system biology approach to identify the influences among COVID-19, influenza, and HIV on the regulation of gene expression.Front Immunol. 2024 Mar 27;15:1369311. doi: 10.3389/fimmu.2024.1369311. eCollection 2024. Front Immunol. 2024. PMID: 38601162 Free PMC article.
-
IFI44 is an immune evasion biomarker for SARS-CoV-2 and Staphylococcus aureus infection in patients with RA.Front Immunol. 2022 Sep 15;13:1013322. doi: 10.3389/fimmu.2022.1013322. eCollection 2022. Front Immunol. 2022. PMID: 36189314 Free PMC article.
-
SARS-CoV-2 and influenza viruses: Strategies to cope with coinfection and bioinformatics perspective.Cell Biol Int. 2022 Jul;46(7):1009-1020. doi: 10.1002/cbin.11800. Epub 2022 Apr 2. Cell Biol Int. 2022. PMID: 35322909 Free PMC article. Review.
Cited by
-
Comparative genomics and integrated system biology approach unveiled undirected phylogeny patterns, mutational hotspots, functional patterns, and molecule repurposing for monkeypox virus.Funct Integr Genomics. 2023 Jul 11;23(3):231. doi: 10.1007/s10142-023-01168-z. Funct Integr Genomics. 2023. PMID: 37432480
References
-
- Abdel-Aziz MI, Kermani NZ, Neerincx AH, Vijverberg SJH, Guo Y, Howarth P, Dahlen SE, Djukanovic R, Sterk PJ, Kraneveld AD, Maitland-van der Zee AH, Chung KF, Adcock IM, On behalf the UBC Association of endopeptidases, involved in SARS-CoV-2 infection, with microbial aggravation in sputum of severe asthma. Allergy. 2021;76:1917–1921. doi: 10.1111/all.14731. - DOI - PubMed
-
- Arguni E, Supriyati E, Hakim MS, Daniwijaya EW, Makrufardi F, Rahayu A, Rovik A, Saraswati U, Oktoviani FN, Prastiwi N, Nuryastuti T, Wibawa T, Haryana SM. Co-infection of SARS-CoV-2 with other viral respiratory pathogens in Yogyakarta, Indonesia: a cross-sectional study. Ann Med Surg (Lond) 2022;77:103676. doi: 10.1016/j.amsu.2022.103676. - DOI - PMC - PubMed
-
- Bai L, Zhao Y, Dong J, Liang S, Guo M, Liu X, Wang X, Huang Z, Sun X, Zhang Z, Dong L, Liu Q, Zheng Y, Niu D, Xiang M, Song K, Ye J, Zheng W, Tang Z, Tang M, Zhou Y, Shen C, Dai M, Zhou L, Chen Y, Yan H, Lan K, Xu K. Coinfection with influenza A virus enhances SARS-CoV-2 infectivity. Cell Res. 2021;31:395–403. doi: 10.1038/s41422-021-00473-1. - DOI - PMC - PubMed
-
- Bankar R, Suvarna K, Ghantasala S, Banerjee A, Biswas D, Choudhury M, Palanivel V, Salkar A, Verma A, Singh A, Mukherjee A, Pai MGJ, Roy J, Srivastava A, Badaya A, Agrawal S, Shrivastav O, Shastri J, Srivastava S. Proteomic investigation reveals dominant alterations of neutrophil degranulation and mRNA translation pathways in patients with COVID-19. iScience. 2021;24:102135. doi: 10.1016/j.isci.2021.102135. - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

