Extensive cytokine biomarker analysis in serum of Guillain-Barré syndrome patients
- PMID: 37221406
- PMCID: PMC10205034
- DOI: 10.1038/s41598-023-35610-w
Extensive cytokine biomarker analysis in serum of Guillain-Barré syndrome patients
Abstract
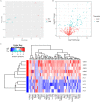
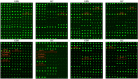
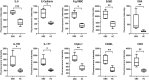
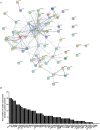
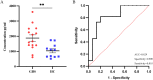
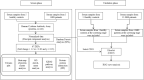
Guillain-Barré syndrome (GBS) is an acute idiopathic polyneuropathy which is related to infection and immune mechanism. The exact pathogenesis of the disease is unknown and treatment is limited. Thus, the purpose of the study is to identify biomarkers of GBS serum and elucidate their involvement in the underlying pathogenesis of GBS that could help to treat GBS more accurately. Antibody array technology was used to detect the expression levels of 440 proteins in serum of 5 GBS group and 5 healthy control group. Sixty-seven differentially expressed proteins (DEPs) were identified by antibody array, among which FoLR1, Legumain, ErbB4, IL-1α, MIP-1α and IGF-2 were down-regulated, while 61 proteins were up-regulated. Bioinformatics analysis indicated that most DEPs were associated with leukocytes, among which IL-1α, SDF-1b, B7-1, CD40, CTLA4, IL-9, MIP-1α and CD40L were in the center of protein-protein interaction (PPI) network. Subsequently, the ability of these DEPs to distinguish GBS from healthy control was further evaluated. CD23 was identified by means of Random Forests Analysis (RFA) and verified by enzyme-linked immunosorbent assay (ELISA). The ROC curve result of CD23 respectively displayed that its sensitivity, specificity and AUC were 0.818, 0.800 and 0.824. We speculate that activation of leukocyte proliferation and migration in circulating blood might be associated with inflammatory recruitment of peripheral nerves, leading to the occurrence and development of GBS, but this conclusion still requires deeper confirmation. More importantly, central proteins may play a pivotal role in the pathogenesis of GBS. In addition, we detected IL-1α, IL-9, and CD23 in the serum of GBS patients for the first time, which may be promising biomarkers for the treatment of GBS.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

