ScRNA-seq revealed an immunosuppression state and tumor microenvironment heterogeneity related to lymph node metastasis in prostate cancer
- PMID: 37221625
- PMCID: PMC10204220
- DOI: 10.1186/s40164-023-00407-0
ScRNA-seq revealed an immunosuppression state and tumor microenvironment heterogeneity related to lymph node metastasis in prostate cancer
Erratum in
-
Correction: ScRNA-seq revealed an immunosuppression state and tumor microenvironment heterogeneity related to lymph node metastasis in prostate cancer.Exp Hematol Oncol. 2024 May 20;13(1):54. doi: 10.1186/s40164-024-00517-3. Exp Hematol Oncol. 2024. PMID: 38769580 Free PMC article. No abstract available.
Abstract
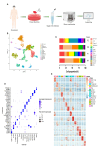
Background: Metastasis is a crucial aspect of disease progression leading to death in patients with prostate cancer (PCa). However, its mechanism remains unclear. We aimed to explore the mechanism of lymph node metastasis (LNM) by analyzing the heterogeneity of tumor microenvironment (TME) in PCa using scRNA-seq.
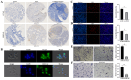
Methods: A total of 32,766 cells were obtained from four PCa tissue samples for scRNA-seq, annotated, and grouped. InferCNV, GSVA, DEG functional enrichment analysis, trajectory analysis, intercellular network evaluation, and transcription factor analysis were carried out for each cell subgroup. Furthermore, validation experiments targeting luminal cell subgroups and CXCR4 + fibroblast subgroup were performed.
Results: The results showed that only EEF2 + and FOLH1 + luminal subgroups were present in LNM, and they appeared at the initial stage of luminal cell differentiation, which were comfirmed by verification experiments. The MYC pathway was enriched in the EEF2 + and FOLH1 + luminal subgroups, and MYC was associated with PCa LNM. Moreover, MYC did not only promote the progression of PCa, but also led to immunosuppression in TME by regulating PDL1 and CD47. The proportion of CD8 + T cells in TME and among NK cells and monocytes was lower in LNM than in the primary lesion, while the opposite was true for Th and Treg cells. Furthermore, these immune cells in TME underwent transcriptional reprogramming, including CD8 + T subgroups of CCR7 + and IL7R+, as well as M2-like monocyte subgroups expressing tumor-associated signature genes, like CCR7, SGKI, and RPL31. Furthermore, STEAP4+, ADGRF5 + and CXCR4+, and SRGNC + fibroblast subgroups were closely related to tumor progression, tumor metabolism, and immunosuppression, indicating their contributions in PCa metastasis. Meanwhile, The presence of CXCR4 + Fibroblasts in PCa was confirmed by polychromatic immunofluorescence.
Conclusions: The significant heterogeneity of luminal, immune, and interstitial cells in PCa LNM may not only directly contribute to tumor progression, but also indirectly result in TME immunosuppression, which may be the cause of metastasis in PCa and in which MYC played an role.
Keywords: Immunosuppression; Metastasis; Prostate Cancer; Single-cell RNA sequencing; Tumor Microenvironment.
© 2023. The Author(s).
Conflict of interest statement
The authors declare that they have no conflicts of interest.
Figures







References
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

