Diffusion tensor imaging-based machine learning for IDH wild-type glioblastoma stratification to reveal the biological underpinning of radiomic features
- PMID: 37222229
- PMCID: PMC10580329
- DOI: 10.1111/cns.14263
Diffusion tensor imaging-based machine learning for IDH wild-type glioblastoma stratification to reveal the biological underpinning of radiomic features
Abstract
Introduction: This study addresses the lack of systematic investigation into the prognostic value of hand-crafted radiomic features derived from diffusion tensor imaging (DTI) in isocitrate dehydrogenase (IDH) wild-type glioblastoma (GBM), as well as the limited understanding of the biological interpretation of individual DTI radiomic features and metrics.
Aims: To develop and validate a DTI-based radiomic model for predicting prognosis in patients with IDH wild-type GBM and reveal the biological underpinning of individual DTI radiomic features and metrics.
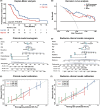
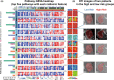
Results: The DTI-based radiomic signature was an independent prognostic factor (p < 0.001). Incorporating the radiomic signature into a clinical model resulted in a radiomic-clinical nomogram that predicted survival better than either the radiomic model or clinical model alone, with a better calibration and classification accuracy. Four categories of pathways (synapse, proliferation, DNA damage response, and complex cellular functions) were significantly correlated with the DTI-based radiomic features and DTI metrics.
Conclusion: The prognostic radiomic features derived from DTI are driven by distinct pathways involved in synapse, proliferation, DNA damage response, and complex cellular functions of GBM.
Keywords: biological pathway; diffusion tensor imaging; glioblastoma; machine learning; prognosis.
© 2023 The Authors. CNS Neuroscience & Therapeutics published by John Wiley & Sons Ltd.
Conflict of interest statement
All authors declare no financial or nonfinancial competing interests.
Figures




References
-
- Hartmann C, Hentschel B, Simon M, et al. Long‐term survival in primary glioblastoma with versus without isocitrate dehydrogenase mutations. Clin Cancer Res. 2013;19(18):5146‐5157. - PubMed
-
- Ramos‐Fresnedo A, Pullen MW, Perez‐Vega C, et al. The survival outcomes of molecular glioblastoma IDH‐wildtype: a multicenter study. J Neurooncol. 2022;157(1):177‐185. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

