Highly sensitive detection of antimicrobial resistance genes in hospital wastewater using the multiplex hybrid capture target enrichment
- PMID: 37222510
- PMCID: PMC10449491
- DOI: 10.1128/msphere.00100-23
Highly sensitive detection of antimicrobial resistance genes in hospital wastewater using the multiplex hybrid capture target enrichment
Abstract
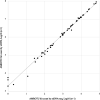
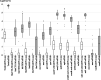
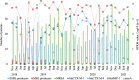
Wastewater can be useful in monitoring the spread of antimicrobial resistance (AMR) within a hospital. The abundance of antibiotic resistance genes (ARGs) in hospital effluent was assessed using metagenomic sequencing (mDNA-seq) and hybrid capture (xHYB). mDNA-seq analysis and subsequent xHYB targeted enrichment were conducted on two effluent samples per month from November 2018 to May 2021. Reads per kilobase per million (RPKM) values were calculated for all 1,272 ARGs in the constructed database. The monthly numbers of patients with presumed extended-spectrum β-lactamase (ESBL)-producing and metallo-β-lactamase (MBL)-producing bacteria, methicillin-resistant Staphylococcus aureus (MRSA), and vancomycin-resistant enterococci (VRE) were compared with the monthly RPKM values of blaCTX-M, blaIMP, mecA, vanA, and vanB by xHYB. The average RPKM value for all ARGs detected by xHYB was significantly higher than that of mDNA-seq (665, 225, and 328, respectively, and P < 0.05). The average number of patients with ESBL producers and RPKM values of blaCTX-M-1 genes in 2020 were significantly higher than that in 2019 (17 and 13 patients per month and 921 vs 232 per month, respectively, both P < 0.05). The average numbers of patients with MBL-producers, MRSA, and VRE were 1, 28, and 0 per month, respectively, while the average RPKM values of blaIMP, mecA, vanA, and vanB were 6,163, 6, 0, and 126 per month, respectively. Monitoring ARGs in hospital effluent using xHYB was found to be more useful than conventional mDNA-seq in detecting ARGs including blaCTX-M, blaIMP, and vanB, which are important for infection control.IMPORTANCEEnvironmental ARGs play a crucial role in the emergence and spread of AMR that constitutes a significant global health threat. One major source of ARGs is effluent from healthcare facilities, where patients are frequently administered antimicrobials. Culture-independent methods, including metagenomics, can detect environmental ARGs carried by non-culturable bacteria and extracellular ARGs. mDNA-seq is one of the most comprehensive methods for environmental ARG surveillance; however, its sensitivity is insufficient for wastewater surveillance. This study demonstrates that xHYB appropriately monitors ARGs in hospital effluent for sensitive identification of nosocomial AMR dissemination. Correlations were observed between the numbers of inpatients with antibiotic-resistant bacteria and the ARG RPKM values in hospital effluent over time. ARG surveillance in hospital effluent using the highly sensitive and specific xHYB method could improve our understanding of the emergence and spread of AMR within a hospital.
Keywords: antibiotic resistant genes; antimicrobial resistance; hospital wastewater; metagenomics; one health; water environment.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
References
-
- Perry MR, Lepper HC, McNally L, Wee BA, Munk P, Warr A, Moore B, Kalima P, Philip C, de Roda Husman AM, Aarestrup FM, Woolhouse MEJ, van Bunnik BAD. 2021. Secrets of the hospital underbelly: patterns of abundance of antimicrobial resistance genes in hospital wastewater vary by specific antimicrobial and bacterial family. Front Microbiol 12: 703560. doi:10.3389/fmicb.2021.703560 - DOI - PMC - PubMed
-
- Beaudry MS, Wang J, Kieran TJ, Thomas J, Bayona-Vásquez NJ, Gao B, Devault A, Brunelle B, Lu K, Wang J-S, Rhodes OE, Glenn TC. 2021. Improved microbial community characterization of 16S rRNA via metagenome hybridization capture enrichment. Front Microbiol 12: 644662. doi:10.3389/fmicb.2021.644662 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

