LncRP11-675F6.3 responds to rapamycin treatment and reduces triglyceride accumulation via interacting with HK1 in hepatocytes by regulating autophagy and VLDL-related proteins
- PMID: 37222534
- PMCID: PMC10577451
- DOI: 10.3724/abbs.2023091
LncRP11-675F6.3 responds to rapamycin treatment and reduces triglyceride accumulation via interacting with HK1 in hepatocytes by regulating autophagy and VLDL-related proteins
Abstract
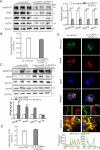
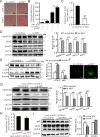
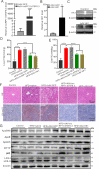
Long noncoding RNAs (lncRNAs) have been widely proven to be involved in liver lipid homeostasis. Herein, we identify an upregulated lncRNA named lncRP11-675F6.3 in response to rapamycin treatment using a microarray in HepG2 cells. Knockdown of lncRP11-675F6. 3 leads to a significant reduction in apolipoprotein 100 (ApoB100), microsomal triglyceride transfer protein (MTTP), ApoE and ApoC3 with increased cellular triglyceride level and autophagy. Furthermore, we find that ApoB100 is obviously colocalized with GFP-LC3 in autophagosomes when lncRP11-675F6. 3 is knocked down, indicating that elevated triglyceride accumulation likely related to autophagy induces the degradation of ApoB100 and impairs very low-density lipoprotein (VLDL) assembly. We then identify and validate that hexokinase 1 (HK1) acts as the binding protein of lncRP11-675F6.3 and mediates triglyceride regulation and cell autophagy. More importantly, we find that lncRP11-675F6.3 and HK1 attenuate high fat diet induced nonalcoholic fatty liver disease (NAFLD) by regulating VLDL-related proteins and autophagy. In conclusion, this study reveals that lncRP11-675F6.3 is potentially involved in the downstream of mTOR signaling pathway and the regulatory network of hepatic triglyceride metabolism in cooperation with its interacting protein HK1, which may provide a new target for fatty liver disorder treatment.
Keywords: autophagy; hexokinase 1; lipoprotein; lncRP11-675F6.3; nonalcoholic fatty liver disease; triglycerides.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures




Similar articles
-
Loss and gain of function of Grp75 or mitofusin 2 distinctly alter cholesterol metabolism, but all promote triglyceride accumulation in hepatocytes.Biochim Biophys Acta Mol Cell Biol Lipids. 2021 Dec;1866(12):159030. doi: 10.1016/j.bbalip.2021.159030. Epub 2021 Aug 20. Biochim Biophys Acta Mol Cell Biol Lipids. 2021. PMID: 34419589
-
The degradation of apolipoprotein B100: multiple opportunities to regulate VLDL triglyceride production by different proteolytic pathways.Biochim Biophys Acta. 2012 May;1821(5):778-81. doi: 10.1016/j.bbalip.2012.02.001. Epub 2012 Feb 10. Biochim Biophys Acta. 2012. PMID: 22342675 Free PMC article. Review.
-
Dysfunction of estrogen-related receptor alpha-dependent hepatic VLDL secretion contributes to sex disparity in NAFLD/NASH development.Theranostics. 2020 Aug 29;10(24):10874-10891. doi: 10.7150/thno.47037. eCollection 2020. Theranostics. 2020. PMID: 33042259 Free PMC article.
-
AUP1 (Ancient Ubiquitous Protein 1) Is a Key Determinant of Hepatic Very-Low-Density Lipoprotein Assembly and Secretion.Arterioscler Thromb Vasc Biol. 2017 Apr;37(4):633-642. doi: 10.1161/ATVBAHA.117.309000. Epub 2017 Feb 9. Arterioscler Thromb Vasc Biol. 2017. PMID: 28183703
-
The ménage à trois of autophagy, lipid droplets and liver disease.Autophagy. 2022 Jan;18(1):50-72. doi: 10.1080/15548627.2021.1895658. Epub 2021 Apr 2. Autophagy. 2022. PMID: 33794741 Free PMC article. Review.
Cited by
-
Targeting LINC070974 inhibits lung adenocarcinoma cell proliferation and progression by interacting with Y-box binding protein 1.Acta Biochim Biophys Sin (Shanghai). 2024 Jun 20;57(2):182-194. doi: 10.3724/abbs.2024093. Acta Biochim Biophys Sin (Shanghai). 2024. PMID: 38899362 Free PMC article.
-
Metabolic Dysfunction-Associated Steatotic Liver Disease: From a Very Low-Density Lipoprotein Perspective.Biomolecules. 2025 Jul 11;15(7):990. doi: 10.3390/biom15070990. Biomolecules. 2025. PMID: 40723862 Free PMC article. Review.
-
ApoB100 and Atherosclerosis: What's New in the 21st Century?Metabolites. 2024 Feb 12;14(2):123. doi: 10.3390/metabo14020123. Metabolites. 2024. PMID: 38393015 Free PMC article. Review.
References
-
- Jones JG. Hepatic glucose and lipid metabolism. Diabetologia. 2016, 59: 1098–1103 - PubMed
-
- Alves-Bezerra M, Cohen DE. Triglyceride metabolism in the liver. Compr Physiol. 2017, 8: 1–8 - PMC - PubMed
-
- Paul B, Lewinska M, Andersen JB. Lipid alterations in chronic liver disease and liver cancer. JHEP reports : innovation in hepatology 2022, 4: 100479 - PMC - PubMed
-
- Younossi ZM, Koenig AB, Abdelatif D, Fazel Y, Henry L, and Wymer M. Global epidemiology of nonalcoholic fatty liver disease-Meta-analytic assessment of prevalence, incidence, and outcomes. Hepatology. (Baltimore, Md.) 2016, 64: 73–84 - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

