Root Niches of Blueberry Imprint Increasing Bacterial-Fungal Interkingdom Interactions along the Soil-Rhizosphere-Root Continuum
- PMID: 37222589
- PMCID: PMC10269492
- DOI: 10.1128/spectrum.05333-22
Root Niches of Blueberry Imprint Increasing Bacterial-Fungal Interkingdom Interactions along the Soil-Rhizosphere-Root Continuum
Abstract
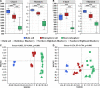
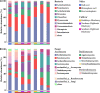
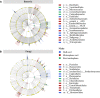
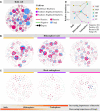
Plant root-associated microbiomes play critical roles in promoting plant health, productivity, and tolerance to biotic/abiotic stresses. Blueberry (Vaccinium spp.) is adapted to acidic soils, while the interactions of the root-associated microbiomes in this specific habitat under various root microenvironments remain elusive. Here, we investigated the diversity and community composition of bacterial and fungal communities in various blueberry root niches (bulk soil, rhizosphere soil, and root endosphere). The results showed that blueberry root niches significantly affected root-associated microbiome diversity and community composition compared to those of the three host cultivars. Deterministic processes gradually increased along the soil-rhizosphere-root continuum in both bacterial and fungal communities. The co-occurrence network topological features showed that both bacterial and fungal community complexity and intensive interactions decreased along the soil-rhizosphere-root continuum. Different compartment niches clearly influenced bacterial-fungal interkingdom interactions, which were significantly higher in the rhizosphere, and positive interactions gradually dominated the co-occurrence networks from the bulk soil to the endosphere. The functional predictions showed that rhizosphere bacterial and fungal communities may have higher cellulolysis and saprotrophy capacities, respectively. Collectively, the root niches not only affected microbial diversity and community composition but also enhanced the positive interkingdom interactions between bacterial and fungal communities along the soil-rhizosphere-root continuum. This provides an essential basis for manipulating synthetic microbial communities for sustainable agriculture. IMPORTANCE The blueberry root-associated microbiome plays an essential role in its adaptation to acidic soils and in limiting the uptake of soil nutrients by its poor root system. Studies on the interactions of the root-associated microbiome in the various root niches may deepen our understanding of the beneficial effects in this particular habitat. Our study extended the research on the diversity and composition of microbial communities in different blueberry root compartment niches. Root niches dominated the root-associated microbiome compared to that of the host cultivar, and deterministic processes increased from the bulk soil to the endosphere. In addition, bacterial-fungal interkingdom interactions were significantly higher in the rhizosphere, and those positive interactions progressively dominated the co-occurrence network along the soil-rhizosphere-root continuum. Collectively, root niches dominantly affected the root-associated microbiome and the positive interkingdom interactions increased, potentially providing benefits for the blueberry.
Keywords: bacterial and fungal communities; co-occurrence networks; compartment niches; rhizosphere; root-associated microbiome.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


Similar articles
-
Host Plant Selection Imprints Structure and Assembly of Fungal Community along the Soil-Root Continuum.mSystems. 2022 Aug 30;7(4):e0036122. doi: 10.1128/msystems.00361-22. Epub 2022 Aug 9. mSystems. 2022. PMID: 35943212 Free PMC article.
-
Ecological Processes of Bacterial and Fungal Communities Associated with Typha orientalis Roots in Wetlands Were Distinct during Plant Development.Microbiol Spectr. 2023 Feb 14;11(1):e0505122. doi: 10.1128/spectrum.05051-22. Epub 2023 Jan 23. Microbiol Spectr. 2023. PMID: 36688664 Free PMC article.
-
Environmental Response to Root Secondary Metabolite Accumulation in Paeonia lactiflora: Insights from Rhizosphere Metabolism and Root-Associated Microbial Communities.Microbiol Spectr. 2022 Dec 21;10(6):e0280022. doi: 10.1128/spectrum.02800-22. Epub 2022 Nov 1. Microbiol Spectr. 2022. PMID: 36318022 Free PMC article.
-
Interkingdom signaling in plant-rhizomicrobiome interactions for sustainable agriculture.Microbiol Res. 2020 Dec;241:126589. doi: 10.1016/j.micres.2020.126589. Epub 2020 Sep 1. Microbiol Res. 2020. PMID: 32927204 Review.
-
Microbiomes in agroecosystem: Diversity, function and assembly mechanisms.Environ Microbiol Rep. 2022 Dec;14(6):833-849. doi: 10.1111/1758-2229.13126. Epub 2022 Oct 2. Environ Microbiol Rep. 2022. PMID: 36184075 Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

