Microbial Assemblages Associated with the Soil-Root Continuum of an Endangered Plant, Helianthemum songaricum Schrenk
- PMID: 37222598
- PMCID: PMC10269481
- DOI: 10.1128/spectrum.03389-22
Microbial Assemblages Associated with the Soil-Root Continuum of an Endangered Plant, Helianthemum songaricum Schrenk
Abstract
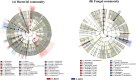
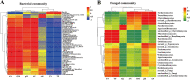
The microbial network of the soil-root continuum plays a key role in plant growth. To date, limited information is available about the microbial assemblages in the rhizosphere and endosphere of endangered plants. We suspect that unknown microorganisms in roots and soil play an important role in the survival strategies of endangered plants. To address this research gap, we investigated the diversity and composition of the microbial communities of the soil-root continuum of the endangered shrub Helianthemum songaricum and observed that the microbial communities and structures of the rhizosphere and endosphere samples were distinguishable. The dominant rhizosphere bacteria were Actinobacteria (36.98%) and Acidobacteria (18.15%), whereas most endophytes were Alphaproteobacteria (23.17%) as well as Actinobacteria (29.94%). The relative abundance of rhizosphere bacteria was higher than that in endosphere samples. Fungal rhizosphere and endophyte samples had approximately equal abundances of the Sordariomycetes (23%), while the Pezizomycetes were more abundant in the soil (31.95%) than in the roots (5.70%). The phylogenetic relationships of the abundances of microbes in root and soil samples also showed that the most abundant bacterial and fungal reads tended to be dominant in either the soil or root samples but not both. Additionally, Pearson correlation heatmap analysis showed that the diversity and composition of soil bacteria and fungi were closely related to pH, total nitrogen, total phosphorus, and organic matter, of which pH and organic matter were the main drivers. These results clarify the different patterns of microbial communities of the soil-root continuum, in support of the better conservation and utilization of endangered desert plants in Inner Mongolia. IMPORTANCE Microbial assemblages play significant roles in plant survival, health, and ecological services. The symbiosis between soil microorganisms and these plants and their interactions with soil factors are important features of the adaptation of desert plants to an arid and barren environment. Therefore, the profound study of the microbial diversity of rare desert plants can provide important data to support the protection and utilization of rare desert plants. Accordingly, in this study, high-throughput sequencing technology was applied to study the microbial diversity in plant roots and rhizosphere soils. We expect that research on the relationship between soil and root microbial diversity and the environment will improve the survival of endangered plants in this environment. In summary, this study is the first to study the microbial diversity and community structure of Helianthemum songaricum Schrenk and compare the diversity and composition of the root and soil microbiomes.
Keywords: Helianthemum songaricum; diversity; endangered plants; high-throughput sequencing; soil-root continuum microbial community.
Conflict of interest statement
The authors declare no conflict of interest.
Figures





Similar articles
-
Epichloë Endophyte Infection Changes the Root Endosphere Microbial Community Composition of Leymus Chinensis Under Both Potted and Field Growth Conditions.Microb Ecol. 2023 Feb;85(2):604-616. doi: 10.1007/s00248-022-01983-0. Epub 2022 Feb 23. Microb Ecol. 2023. PMID: 35194659
-
Comparison of network connectivity and environmental driving factors of root-associated fungal communities of desert ephemeral plants in two habitat soils.J Environ Manage. 2023 Apr 15;332:117375. doi: 10.1016/j.jenvman.2023.117375. Epub 2023 Jan 28. J Environ Manage. 2023. PMID: 36716547
-
Distinct Bacterial Communities Within the Nonrhizosphere, Rhizosphere, and Endosphere of Ammodendron bifolium Under Winter Condition in the Takeermohuer Desert.Microb Ecol. 2024 Nov 29;87(1):151. doi: 10.1007/s00248-024-02462-4. Microb Ecol. 2024. PMID: 39611982 Free PMC article.
-
[Mechanisms of Rhizosphere Microorganisms in Regulating Plant Root System Architecture in Acidic Soils].Huan Jing Ke Xue. 2025 Jan 8;46(1):570-578. doi: 10.13227/j.hjkx.202312260. Huan Jing Ke Xue. 2025. PMID: 39721662 Review. Chinese.
-
New Aspects of the Effects of Climate Change on Interactions Between Plants and Microbiomes: A Review.J Basic Microbiol. 2024 Oct;64(10):e2400345. doi: 10.1002/jobm.202400345. Epub 2024 Aug 28. J Basic Microbiol. 2024. PMID: 39205430 Review.
Cited by
-
Comparative analysis of actinorhizal nodule and associated soil microorganism diversity and structure in three Alnus species.Front Plant Sci. 2025 May 8;16:1572494. doi: 10.3389/fpls.2025.1572494. eCollection 2025. Front Plant Sci. 2025. PMID: 40406724 Free PMC article.
-
Characterizing the microbiome recruited by the endangered plant Firmiana danxiaensis in phosphorus-deficient acidic soil.Front Microbiol. 2025 Jan 15;15:1439446. doi: 10.3389/fmicb.2024.1439446. eCollection 2024. Front Microbiol. 2025. PMID: 39881984 Free PMC article.
References
-
- Kang H, Yu W, Dutta S, Gao H. 2021. Soil microbial community composition and function are closely associated with soil organic matter chemistry along a latitudinal gradient. Geoderma 383:114744. doi:10.1016/j.geoderma.2020.114744. - DOI
-
- Sneha GR, Swarnalakshmi K, Sharma M, Reddy K, Bhoumik A, Suman A, Kannepalli A. 2021. Soil type influence nutrient availability, microbial metabolic diversity, eubacterial and diazotroph abundance in chickpea rhizosphere. World J Microbiol Biotechnol 37:167. doi:10.1007/s11274-021-03132-0. - DOI - PubMed
-
- Gao F, Fan H, Chapman SJ, Yao H. 2022. Changes in soil microbial community activity and composition following substrate amendment within the MicroResp system. J Soils Sediments 22:1242–1251. doi:10.1007/s11368-022-03143-w. - DOI
-
- Wang J, Liao L, Ye Z, Liu H, Zhang C, Zhang L, Liu G, Wang G. 2022. Different bacterial co-occurrence patterns and community assembly between rhizosphere and bulk soils under N addition in the plant-soil system. Plant Soil 471:697–713. doi:10.1007/s11104-021-05214-2. - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

