A leader cell triggers end of lag phase in populations of Pseudomonas fluorescens
- PMID: 37223352
- PMCID: PMC10117806
- DOI: 10.1093/femsml/uqac022
A leader cell triggers end of lag phase in populations of Pseudomonas fluorescens
Erratum in
-
Correction to: A leader cell triggers end of lag phase in populations of Pseudomonas fluorescens.Microlife. 2023 Oct 20;4:uqad040. doi: 10.1093/femsml/uqad040. eCollection 2023. Microlife. 2023. PMID: 37869483 Free PMC article.
Abstract
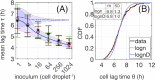
The relationship between the number of cells colonizing a new environment and time for resumption of growth is a subject of long-standing interest. In microbiology this is known as the "inoculum effect." Its mechanistic basis is unclear with possible explanations ranging from the independent actions of individual cells, to collective actions of populations of cells. Here, we use a millifluidic droplet device in which the growth dynamics of hundreds of populations founded by controlled numbers of Pseudomonas fluorescens cells, ranging from a single cell, to one thousand cells, were followed in real time. Our data show that lag phase decreases with inoculum size. The decrease of average lag time and its variance across droplets, as well as lag time distribution shapes, follow predictions of extreme value theory, where the inoculum lag time is determined by the minimum value sampled from the single-cell distribution. Our experimental results show that exit from lag phase depends on strong interactions among cells, consistent with a "leader cell" triggering end of lag phase for the entire population.
Keywords: collective behavior; extreme value theory; growth dynamics; high-throughput millifluidics; microbial population biology.
© The Author(s) 2022. Published by Oxford University Press on behalf of FEMS.
Conflict of interest statement
No competing interests are declared.
Figures



Comment in
-
Correction to: A leader cell triggers end of lag phase in populations of Pseudomonas fluorescens.Microlife. 2023 Oct 20;4:uqad040. doi: 10.1093/femsml/uqad040. eCollection 2023. Microlife. 2023. PMID: 37869483 Free PMC article.
References
-
- Baraban L, Bertholle F, Salverda MLMet al. Millifluidic droplet analyser for microbiology. Lab Chip. 2011;11:4057–62. - PubMed
LinkOut - more resources
Full Text Sources
