Nonessential tRNA and rRNA modifications impact the bacterial response to sub-MIC antibiotic stress
- PMID: 37223353
- PMCID: PMC10117853
- DOI: 10.1093/femsml/uqac019
Nonessential tRNA and rRNA modifications impact the bacterial response to sub-MIC antibiotic stress
Abstract
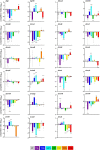
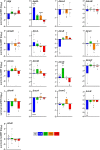
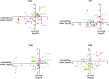
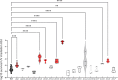
Antimicrobial resistance develops as a major problem in infectious diseases treatment. While antibiotic resistance mechanisms are usually studied using lethal antibiotic doses, lower doses allowing bacterial growth are now considered as factors influencing the development and selection of resistance. Starting with a high-density Tn insertion library in Vibrio cholerae and following its evolution by TN-seq in the presence of subinhibitory concentrations of antibiotics, we discovered that RNA modification genes can have opposite fates, being selected or counter-selected. We, thus have undertaken the phenotypic characterization of 23 transfer RNA (tRNA) and ribosomal RNA (rRNA) modifications deletion mutants, for which growth is globally not affected in the absence of stress. We uncover a specific involvement of different RNA modification genes in the response to aminoglycosides (tobramycin and gentamicin), fluoroquinolones (ciprofloxacin), β-lactams (carbenicillin), chloramphenicol, and trimethoprim. Our results identify t/rRNA modification genes, not previously associated to any antibiotic resistance phenotype, as important factors affecting the bacterial response to low doses of antibiotics from different families. This suggests differential translation and codon decoding as critical factors involved in the bacterial response to stress.
Keywords: RNA modifications; Vibrio cholerae; antibiotic resistance; bacterial stress responses; differential translation; sub-MIC antibiotics.
© The Author(s) 2022. Published by Oxford University Press on behalf of FEMS.
Conflict of interest statement
None declared.
Figures


References
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
