Bacterial methyltransferases: from targeting bacterial genomes to host epigenetics
- PMID: 37223361
- PMCID: PMC10117894
- DOI: 10.1093/femsml/uqac014
Bacterial methyltransferases: from targeting bacterial genomes to host epigenetics
Abstract
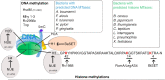
Methyltransferase (MTases) enzymes transfer methyl groups particularly on proteins and nucleotides, thereby participating in controlling the epigenetic information in both prokaryotes and eukaryotes. The concept of epigenetic regulation by DNA methylation has been extensively described for eukaryotes. However, recent studies have extended this concept to bacteria showing that DNA methylation can also exert epigenetic control on bacterial phenotypes. Indeed, the addition of epigenetic information to nucleotide sequences confers adaptive traits including virulence-related characteristics to bacterial cells. In eukaryotes, an additional layer of epigenetic regulation is obtained by post-translational modifications of histone proteins. Interestingly, in the last decades it was shown that bacterial MTases, besides playing an important role in epigenetic regulations at the microbe level by exerting an epigenetic control on their own gene expression, are also important players in host-microbe interactions. Indeed, secreted nucleomodulins, bacterial effectors that target the nucleus of infected cells, have been shown to directly modify the epigenetic landscape of the host. A subclass of nucleomodulins encodes MTase activities, targeting both host DNA and histone proteins, leading to important transcriptional changes in the host cell. In this review, we will focus on lysine and arginine MTases of bacteria and their hosts. The identification and characterization of these enzymes will help to fight bacterial pathogens as they may emerge as promising targets for the development of novel epigenetic inhibitors in both bacteria and the host cells they infect.
Keywords: Legionella; bacterial pathogens; epigenetics; methyltransferase.
© The Author(s) 2021. Published by Oxford University Press on behalf of FEMS.
Conflict of interest statement
The authors declare no conflict of interest
Figures


Similar articles
-
Bacterial DNA methyltransferase: A key to the epigenetic world with lessons learned from proteobacteria.Front Microbiol. 2023 Mar 22;14:1129437. doi: 10.3389/fmicb.2023.1129437. eCollection 2023. Front Microbiol. 2023. PMID: 37032876 Free PMC article. Review.
-
Bacterial Factors Targeting the Nucleus: The Growing Family of Nucleomodulins.Toxins (Basel). 2020 Mar 31;12(4):220. doi: 10.3390/toxins12040220. Toxins (Basel). 2020. PMID: 32244550 Free PMC article. Review.
-
Chemical Expansion of the Methyltransferase Reaction: Tools for DNA Labeling and Epigenome Analysis.Acc Chem Res. 2023 Nov 21;56(22):3188-3197. doi: 10.1021/acs.accounts.3c00471. Epub 2023 Oct 30. Acc Chem Res. 2023. PMID: 37904501 Free PMC article.
-
Bacterial nucleomodulins: A coevolutionary adaptation to the eukaryotic command center.PLoS Pathog. 2021 Jan 21;17(1):e1009184. doi: 10.1371/journal.ppat.1009184. eCollection 2021 Jan. PLoS Pathog. 2021. PMID: 33476322 Free PMC article. Review.
-
DNA Methyltransferase Regulates Nitric Oxide Homeostasis and Virulence in a Chronically Adapted Pseudomonas aeruginosa Strain.mSystems. 2022 Oct 26;7(5):e0043422. doi: 10.1128/msystems.00434-22. Epub 2022 Sep 15. mSystems. 2022. PMID: 36106744 Free PMC article.
Cited by
-
Moving toward the Inclusion of Epigenomics in Bacterial Genome Evolution: Perspectives and Challenges.Int J Mol Sci. 2024 Apr 17;25(8):4425. doi: 10.3390/ijms25084425. Int J Mol Sci. 2024. PMID: 38674013 Free PMC article. Review.
-
Dual RNA-Seq of Flavobacterium psychrophilum and Its Outer Membrane Vesicles Distinguishes Genes Associated with Susceptibility to Bacterial Cold-Water Disease in Rainbow Trout (Oncorhynchus mykiss).Pathogens. 2023 Mar 10;12(3):436. doi: 10.3390/pathogens12030436. Pathogens. 2023. PMID: 36986358 Free PMC article.
-
Single-base resolution quantitative genome methylation analysis in the model bacterium Helicobacter pylori by enzymatic methyl sequencing (EM-Seq) reveals influence of strain, growth phase, and methyl homeostasis.BMC Biol. 2024 May 29;22(1):125. doi: 10.1186/s12915-024-01921-1. BMC Biol. 2024. PMID: 38807090 Free PMC article.
-
Surviving Colonies of Pseudomonas aeruginosa Isolated In Vivo from Infected, Antibiotic-Treated Galleria mellonella Larvae Acquire an Antibiotic-Tolerant Phenotype.Antibiotics (Basel). 2025 May 15;14(5):507. doi: 10.3390/antibiotics14050507. Antibiotics (Basel). 2025. PMID: 40426573 Free PMC article.
-
Legionella pneumophila modulates macrophage functions through epigenetic reprogramming via the C-type lectin receptor Mincle.iScience. 2024 Aug 8;27(9):110700. doi: 10.1016/j.isci.2024.110700. eCollection 2024 Sep 20. iScience. 2024. PMID: 39252966 Free PMC article.
References
-
- Adhikari S, Curtis PD. DNA methyltransferases and epigenetic regulation in bacteria. FEMS Microbiol Rev. 2016;40:575–91. - PubMed
Publication types
LinkOut - more resources
Full Text Sources
