Mass spectrometry-based assays for assessing replicative bypass and repair of DNA alkylation in cells
- PMID: 37223415
- PMCID: PMC10201546
- DOI: 10.1039/d2ra08340j
Mass spectrometry-based assays for assessing replicative bypass and repair of DNA alkylation in cells
Abstract
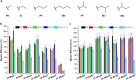
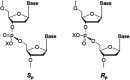
Endogenous metabolism and environmental exposure can give rise to DNA alkylation, which can elicit deleterious biological consequences. In the search for reliable and quantitative analytical methods to elucidate the impact of DNA alkylation on the flow of genetic information, mass spectrometry (MS) has attracted increasing attention, owing to its unambiguous determination of molecular mass. The MS-based assays obviate conventional colony-picking methods and Sanger sequencing procedures, and retained the high sensitivity of postlabeling methods. With the help of the CRISPR/Cas9 gene editing method, MS-based assays showed high potential in studying individual functions of repair proteins and translesion synthesis (TLS) polymerases in DNA replication. In this mini-review, we have summarized the development of MS-based competitive and replicative adduct bypass (CRAB) assays and their recent applications in assessing the impact of alkylation on DNA replication. With further development of MS instruments for high resolving power and high throughput, these assays should be generally applicable and efficient in quantitative measurement of the biological consequences and repair of other DNA lesions.
This journal is © The Royal Society of Chemistry.
Conflict of interest statement
There are no conflicts to declare.
Figures








Similar articles
-
Mass Spectrometry-Based Quantitative Strategies for Assessing the Biological Consequences and Repair of DNA Adducts.Acc Chem Res. 2016 Feb 16;49(2):205-13. doi: 10.1021/acs.accounts.5b00437. Epub 2016 Jan 13. Acc Chem Res. 2016. PMID: 26758048 Free PMC article. Review.
-
Cytotoxic and mutagenic properties of minor-groove O2-alkylthymidine lesions in human cells.J Biol Chem. 2018 Jun 1;293(22):8638-8644. doi: 10.1074/jbc.RA118.003133. Epub 2018 Apr 23. J Biol Chem. 2018. PMID: 29685891 Free PMC article.
-
Translesion synthesis of O4-alkylthymidine lesions in human cells.Nucleic Acids Res. 2016 Nov 2;44(19):9256-9265. doi: 10.1093/nar/gkw662. Epub 2016 Jul 27. Nucleic Acids Res. 2016. PMID: 27466394 Free PMC article.
-
Impact of tobacco-specific nitrosamine-derived DNA adducts on the efficiency and fidelity of DNA replication in human cells.J Biol Chem. 2018 Jul 13;293(28):11100-11108. doi: 10.1074/jbc.RA118.003477. Epub 2018 May 22. J Biol Chem. 2018. PMID: 29789427 Free PMC article.
-
Family A and B DNA Polymerases in Cancer: Opportunities for Therapeutic Interventions.Biology (Basel). 2018 Jan 2;7(1):5. doi: 10.3390/biology7010005. Biology (Basel). 2018. PMID: 29301327 Free PMC article. Review.
References
-
- Friedberg E. C., Walker G. C., Siede W., Wood R. D., Schultz R. A. and Ellenberger T., DNA Repair and Mutagenesis, ASM Press, Washington, D.C., 2005
Publication types
LinkOut - more resources
Full Text Sources

