The essential role of N6-methyladenosine RNA methylation in complex eye diseases
- PMID: 37223523
- PMCID: PMC10201676
- DOI: 10.1016/j.gendis.2022.05.008
The essential role of N6-methyladenosine RNA methylation in complex eye diseases
Abstract
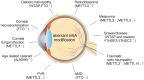
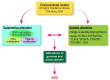
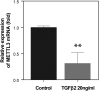
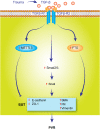
There are many complex eye diseases which are the leading causes of blindness, however, the pathogenesis of the complex eye diseases is not fully understood, especially the underlying molecular mechanisms of N6-methyladenosine (m6A) RNA methylation in the eye diseases have not been extensive clarified. Our review summarizes the latest advances in the studies of m6A modification in the pathogenesis of the complex eye diseases, including cornea disease, cataract, diabetic retinopathy, age-related macular degeneration, proliferative vitreoretinopathy, Graves' disease, uveal melanoma, retinoblastoma, and traumatic optic neuropathy. We further discuss the possibility of developing m6A modification signatures as biomarkers for the diagnosis of the eye diseases, as well as potential therapeutic approaches.
Keywords: Degeneration; Eye diseases; Fibrosis; Inflammation; Tumor; m6A RNA methylation.
© 2022 The Authors. Publishing services by Elsevier B.V. on behalf of KeAi Communications Co., Ltd.
Figures


Similar articles
-
Role of m6A methylation in retinal diseases.Exp Eye Res. 2023 Jun;231:109489. doi: 10.1016/j.exer.2023.109489. Epub 2023 Apr 20. Exp Eye Res. 2023. PMID: 37084873 Review.
-
Potential Impact of N6-Methyladenosine RNA Methylation on Vision Function and the Pathological Processes of Ocular Diseases: New Discoveries and Future Perspectives.Front Biosci (Landmark Ed). 2022 Jun 28;27(7):207. doi: 10.31083/j.fbl2707207. Front Biosci (Landmark Ed). 2022. PMID: 35866387 Review.
-
N6-methyladenosine methylation in ophthalmic diseases: From mechanisms to potential applications.Heliyon. 2023 Dec 13;10(1):e23668. doi: 10.1016/j.heliyon.2023.e23668. eCollection 2024 Jan 15. Heliyon. 2023. PMID: 38192819 Free PMC article. Review.
-
The role of N6-methyladenosine (m6A) in eye diseases.Mol Biol Rep. 2021 Aug;48(8):6145-6150. doi: 10.1007/s11033-021-06596-3. Epub 2021 Jul 31. Mol Biol Rep. 2021. PMID: 34331665 Review.
-
N6-methyladenosine (m6A) methylation in kidney diseases: Mechanisms and therapeutic potential.Biochim Biophys Acta Gene Regul Mech. 2023 Dec;1866(4):194967. doi: 10.1016/j.bbagrm.2023.194967. Epub 2023 Aug 6. Biochim Biophys Acta Gene Regul Mech. 2023. PMID: 37553065 Review.
Cited by
-
Interpretability-guided RNA N6-methyladenosine modification site prediction with invertible neural networks.Commun Biol. 2025 Jul 8;8(1):1022. doi: 10.1038/s42003-025-08265-8. Commun Biol. 2025. PMID: 40629144 Free PMC article.
-
Evaluation of Gene Expression and the Regulatory Role of microRNAs Related to the Mitogen-Activated Protein Kinase Signaling Pathway in Human Retinal Pigment Epithelial Cells Treated With Lipopolysaccharide A and Tacrolimus.Mediators Inflamm. 2025 Jul 1;2025:8586711. doi: 10.1155/mi/8586711. eCollection 2025. Mediators Inflamm. 2025. PMID: 40630080 Free PMC article.
-
WTAP-mediated N6-methyladenosine mRNA methylation regulates laser-induced macular neovascularization.Mol Vis. 2024 Oct 8;30:336-347. eCollection 2024. Mol Vis. 2024. PMID: 40330498 Free PMC article.
-
LncRNA HOTAIR Interaction With WTAP Promotes m6A Methyltransferase Complex Assembly and Posterior Capsule Opacification Formation by Increasing THBS1.Invest Ophthalmol Vis Sci. 2025 May 1;66(5):20. doi: 10.1167/iovs.66.5.20. Invest Ophthalmol Vis Sci. 2025. PMID: 40341312 Free PMC article.
-
METTL3-mediated m6A modification of SIRT1 mRNA affects the progression of diabetic cataracts through cellular autophagy and senescence.J Transl Med. 2024 Sep 27;22(1):865. doi: 10.1186/s12967-024-05691-w. J Transl Med. 2024. PMID: 39334185 Free PMC article.
References
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources

