Structural and functional diversity of bacterial cyclic nucleotide perception by CRP proteins
- PMID: 37223727
- PMCID: PMC10187061
- DOI: 10.1093/femsml/uqad024
Structural and functional diversity of bacterial cyclic nucleotide perception by CRP proteins
Abstract
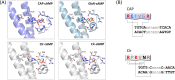
Cyclic AMP (cAMP) is a ubiquitous second messenger synthesized by most living organisms. In bacteria, it plays highly diverse roles in metabolism, host colonization, motility, and many other processes important for optimal fitness. The main route of cAMP perception is through transcription factors from the diverse and versatile CRP-FNR protein superfamily. Since the discovery of the very first CRP protein CAP in Escherichia coli more than four decades ago, its homologs have been characterized in both closely related and distant bacterial species. The cAMP-mediated gene activation for carbon catabolism by a CRP protein in the absence of glucose seems to be restricted to E. coli and its close relatives. In other phyla, the regulatory targets are more diverse. In addition to cAMP, cGMP has recently been identified as a ligand of certain CRP proteins. In a CRP dimer, each of the two cyclic nucleotide molecules makes contacts with both protein subunits and effectuates a conformational change that favors DNA binding. Here, we summarize the current knowledge on structural and physiological aspects of E. coli CAP compared with other cAMP- and cGMP-activated transcription factors, and point to emerging trends in metabolic regulation related to lysine modification and membrane association of CRP proteins.
Keywords: CRP regulon; CRP transcription factors; allosteric control; cAMP; cGMP; nucleotide second messenger.
© The Author(s) 2023. Published by Oxford University Press on behalf of FEMS.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
Publication types
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous
