Fine mapping of a QTL and identification of candidate genes associated with cold tolerance during germination in peanut (Arachis hypogaea L.) on chromosome B09 using whole genome re-sequencing
- PMID: 37223785
- PMCID: PMC10200878
- DOI: 10.3389/fpls.2023.1153293
Fine mapping of a QTL and identification of candidate genes associated with cold tolerance during germination in peanut (Arachis hypogaea L.) on chromosome B09 using whole genome re-sequencing
Abstract
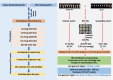
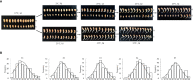
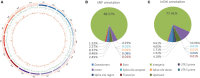
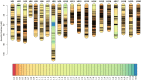
Low temperatures significantly affect the growth and yield of peanuts. Temperatures lower than 12 °C are generally detrimental for the germination of peanuts. To date, there has been no report on precise information on the quantitative trait loci (QTL) for cold tolerance during the germination in peanuts. In this study, we developed a recombinant inbred line (RIL) population comprising 807 RILs by tolerant and sensitive parents. Phenotypic frequencies of germination rate low-temperature conditions among RIL population showed normally distributed in five environments. Then, we constructed a high density SNP-based genetic linkage map through whole genome re-sequencing (WGRS) technique and identified a major quantitative trait locus (QTL), qRGRB09, on chromosome B09. The cold tolerance-related QTLs were repeatedly detected in all five environments, and the genetic distance was 6.01 cM (46.74 cM - 61.75 cM) after taking a union set. To further confirm that qRGRB09 was located on chromosome B09, we developed Kompetitive Allele Specific PCR (KASP) markers for the corresponding QTL regions. A regional QTL mapping analysis, which was conducted after taking the intersection of QTL intervals of all environments into account, confirmed that qRGRB09 was between the KASP markers, G22096 and G220967 (chrB09:155637831-155854093), and this region was 216.26 kb in size, wherein a total of 15 annotated genes were detected. This study illustrates the relevance of WGRS-based genetic maps for QTL mapping and KASP genotyping that facilitated QTL fine mapping of peanuts. The results of our study also provided useful information on the genetic architecture underlying cold tolerance during germination in peanuts, which in turn may be useful for those engaged in molecular studies as well as crop improvement in the cold-stressed environment.
Keywords: QTL; candidate genes; cold tolerance; germination; peanut; whole genome re-sequencing.
Copyright © 2023 Zhang, Zhang, Wang, Liu, Liang, Zhang, Xue, Tian, Zhang, Li, Sheng, Nie, Feng, Liao and Bai.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


References
-
- Agarwal G., Clevenger J., Pandey M. K., Wang H., Shasidhar Y., Chu Y., et al. (2018). High-density genetic map using whole-genome resequencing for fine mapping and candidate gene discovery for disease resistance in peanut. Plant Biotechnol. J. 16 (11), 1954–1967. doi: 10.1111/pbi.12930 - DOI - PMC - PubMed
-
- Bai D. M., Xue Y. Y., Huang L., Huai D. X., Tian Y. X., Wang P. D., et al. (2022). Assessment of cold tolerance of different peanut varieties and screening of evaluation indexes at germination stage. Acta Agronomica Sin. 48 (8), 2066–2079. doi: 10.3724/SP.J.1006.2022.14163 - DOI
-
- Bai D. M., Xue Y. Y., Zhao J. J., Huang L., Jiang H. F. (2018). Identification of cold-tolerance during germination stage and genetic diversity of SSR markers in peanut landraces of shanxi province. Acta Agronomica Sin. 44 (10), 1459. doi: 10.3724/SP.J.1006.2018.01459 - DOI
LinkOut - more resources
Full Text Sources

