Drought-triggered leaf transcriptional responses disclose key molecular pathways underlying leaf water use efficiency in sugarcane (Saccharum spp.)
- PMID: 37223790
- PMCID: PMC10200899
- DOI: 10.3389/fpls.2023.1182461
Drought-triggered leaf transcriptional responses disclose key molecular pathways underlying leaf water use efficiency in sugarcane (Saccharum spp.)
Abstract
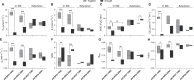
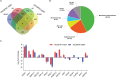
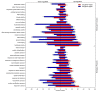
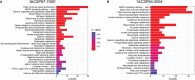
Drought is a major constraint to sugarcane (Saccharum spp.) production and improving the water use efficiency (WUE) is a critical trait for the sustainability of this bioenergy crop. The molecular mechanism underlying WUE remains underexplored in sugarcane. Here, we investigated the drought-triggered physiological and transcriptional responses of two sugarcane cultivars contrasting for drought tolerance, 'IACSP97-7065' (sensitive) and 'IACSP94-2094' (tolerant). After 21 days without irrigation (DWI), only 'IACSP94-2094' exhibited superior WUE and instantaneous carboxylation efficiency, with the net CO2 assimilation being less impacted when compared with 'IACSP97-7065'. RNA-seq of sugarcane leaves at 21 DWI revealed a total of 1,585 differentially expressed genes (DEGs) for both genotypes, among which 'IACSP94-2094' showed 617 (38.9%) exclusive transcripts (212 up- and 405 down-regulated). Functional enrichment analyses of these unique DEGs revealed several relevant biological processes, such as photosynthesis, transcription factors, signal transduction, solute transport, and redox homeostasis. The better drought-responsiveness of 'IACSP94-2094' suggested signaling cascades that foster transcriptional regulation of genes implicated in the Calvin cycle and transport of water and carbon dioxide, which are expected to support the high WUE and carboxylation efficiency observed for this genotype under water deficit. Moreover, the robust antioxidant system of the drought-tolerant genotype might serve as a molecular shield against the drought-associated overproduction of reactive oxygen species. This study provides relevant data that may be used to develop novel strategies for sugarcane breeding programs and to understand the genetic basis of drought tolerance and WUE improvement of sugarcane.
Keywords: abiotic stress; antioxidant mechanism; carboxylation efficiency; transcriptome; water use efficiency.
Copyright © 2023 Contiliani, Nebó, Ribeiro, Landell, Pereira, Ming, Figueira and Creste.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

Similar articles
-
Leaf transcriptome profiling of contrasting sugarcane genotypes for drought tolerance under field conditions.Sci Rep. 2022 Jun 1;12(1):9153. doi: 10.1038/s41598-022-13158-5. Sci Rep. 2022. PMID: 35650424 Free PMC article.
-
Short-term physiological changes in roots and leaves of sugarcane varieties exposed to H2O2 in root medium.J Plant Physiol. 2015 Apr 1;177:93-99. doi: 10.1016/j.jplph.2015.01.009. Epub 2015 Jan 25. J Plant Physiol. 2015. PMID: 25703773
-
Superoxide dismutase and ascorbate peroxidase improve the recovery of photosynthesis in sugarcane plants subjected to water deficit and low substrate temperature.Plant Physiol Biochem. 2013 Dec;73:326-36. doi: 10.1016/j.plaphy.2013.10.012. Epub 2013 Oct 17. Plant Physiol Biochem. 2013. PMID: 24184453
-
MicroRNAs and drought responses in sugarcane.Front Plant Sci. 2015 Feb 23;6:58. doi: 10.3389/fpls.2015.00058. eCollection 2015. Front Plant Sci. 2015. PMID: 25755657 Free PMC article. Review.
-
Drought and salinity stresses induced physio-biochemical changes in sugarcane: an overview of tolerance mechanism and mitigating approaches.Front Plant Sci. 2023 Aug 11;14:1225234. doi: 10.3389/fpls.2023.1225234. eCollection 2023. Front Plant Sci. 2023. PMID: 37645467 Free PMC article. Review.
References
-
- BioBam Bioinformatics (2019) OmicsBox - bioinformatics made easy. Available at: https://www.biobam.com/omicsbox (Accessed February 16, 2023).
LinkOut - more resources
Full Text Sources
Miscellaneous

