Design and Selection of Heterodimerizing Helical Hairpins for Synthetic Biology
- PMID: 37224449
- PMCID: PMC10278171
- DOI: 10.1021/acssynbio.3c00231
Design and Selection of Heterodimerizing Helical Hairpins for Synthetic Biology
Abstract
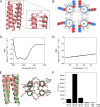
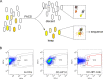
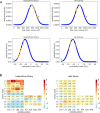
Synthetic biology applications would benefit from protein modules of reduced complexity that function orthogonally to cellular components. As many subcellular processes depend on peptide-protein or protein-protein interactions, de novo designed polypeptides that can bring together other proteins controllably are particularly useful. Thanks to established sequence-to-structure relationships, helical bundles provide good starting points for such designs. Typically, however, such designs are tested in vitro and function in cells is not guaranteed. Here, we describe the design, characterization, and application of de novo helical hairpins that heterodimerize to form 4-helix bundles in cells. Starting from a rationally designed homodimer, we construct a library of helical hairpins and identify complementary pairs using bimolecular fluorescence complementation in E. coli. We characterize some of the pairs using biophysics and X-ray crystallography to confirm heterodimeric 4-helix bundles. Finally, we demonstrate the function of an exemplar pair in regulating transcription in both E. coli and mammalian cells.
Keywords: coiled coil; in-cell library screening; protein−protein interactions; rational peptide design; synthetic biology.
Conflict of interest statement
The authors declare no competing financial interest.
Figures


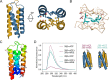
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

