Deep integrative models for large-scale human genomics
- PMID: 37224538
- PMCID: PMC10325897
- DOI: 10.1093/nar/gkad373
Deep integrative models for large-scale human genomics
Abstract
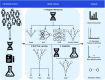
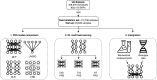
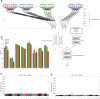
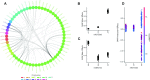
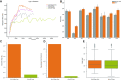
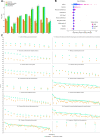
Polygenic risk scores (PRSs) are expected to play a critical role in precision medicine. Currently, PRS predictors are generally based on linear models using summary statistics, and more recently individual-level data. However, these predictors mainly capture additive relationships and are limited in data modalities they can use. We developed a deep learning framework (EIR) for PRS prediction which includes a model, genome-local-net (GLN), specifically designed for large-scale genomics data. The framework supports multi-task learning, automatic integration of other clinical and biochemical data, and model explainability. When applied to individual-level data from the UK Biobank, the GLN model demonstrated a competitive performance compared to established neural network architectures, particularly for certain traits, showcasing its potential in modeling complex genetic relationships. Furthermore, the GLN model outperformed linear PRS methods for Type 1 Diabetes, likely due to modeling non-additive genetic effects and epistasis. This was supported by our identification of widespread non-additive genetic effects and epistasis in the context of T1D. Finally, we constructed PRS models that integrated genotype, blood, urine, and anthropometric data and found that this improved performance for 93% of the 290 diseases and disorders considered. EIR is available at https://github.com/arnor-sigurdsson/EIR.
© The Author(s) 2023. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures
References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

