Cross-tissue omics analysis discovers ten adipose genes encoding secreted proteins in obesity-related non-alcoholic fatty liver disease
- PMID: 37224770
- PMCID: PMC10277924
- DOI: 10.1016/j.ebiom.2023.104620
Cross-tissue omics analysis discovers ten adipose genes encoding secreted proteins in obesity-related non-alcoholic fatty liver disease
Abstract
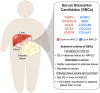
Background: Non-alcoholic fatty liver disease (NAFLD) is a fast-growing, underdiagnosed, epidemic. We hypothesise that obesity-related inflammation compromises adipose tissue functions, preventing efficient fat storage, and thus driving ectopic fat accumulation into the liver.
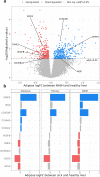
Methods: To identify adipose-based mechanisms and potential serum biomarker candidates (SBCs) for NAFLD, we utilise dual-tissue RNA-sequencing (RNA-seq) data in adipose tissue and liver, paired with histology-based NAFLD diagnosis, from the same individuals in a cohort of obese individuals. We first scan for genes that are differentially expressed (DE) for NAFLD in obese individuals' subcutaneous adipose tissue but not in their liver; encode proteins secreted to serum; and show preferential adipose expression. Then the identified genes are filtered to key adipose-origin NAFLD genes by best subset analysis, knockdown experiments during human preadipocyte differentiation, recombinant protein treatment experiments in human liver HepG2 cells, and genetic analysis.
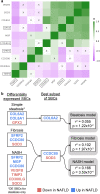
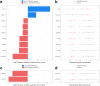
Findings: We discover a set of genes, including 10 SBCs, that may modulate NAFLD pathogenesis by impacting adipose tissue function. Based on best subset analysis, we further follow-up on two SBCs CCDC80 and SOD3 by knockdown in human preadipocytes and subsequent differentiation experiments, which show that they modulate crucial adipogenesis genes, LPL, SREBPF1, and LEP. We also show that treatment of the liver HepG2 cells with the CCDC80 and SOD3 recombinant proteins impacts genes related to steatosis and lipid processing, including PPARA, NFE2L2, and RNF128. Finally, utilizing the adipose NAFLD DE gene cis-regulatory variants associated with serum triglycerides (TGs) in extensive genome-wide association studies (GWASs), we demonstrate a unidirectional effect of serum TGs on NAFLD with Mendelian Randomization (MR) analysis. We also demonstrate that a single SNP regulating one of the SBC genes, rs2845885, produces a significant MR result by itself. This supports the conclusion that genetically regulated adipose expression of the NAFLD DE genes may contribute to NAFLD through changes in serum TG levels.
Interpretation: Our results from the dual-tissue transcriptomics screening improve the understanding of obesity-related NAFLD by providing a targeted set of 10 adipose tissue-active genes as new serum biomarker candidates for the currently grossly underdiagnosed fatty liver disease.
Funding: The work was supported by NIH grants R01HG010505 and R01DK132775. The Genotype-Tissue Expression (GTEx) Project was supported by the Common Fund of the Office of the Director of the National Institutes of Health, and by NCI, NHGRI, NHLBI, NIDA, NIMH, and NINDS. The KOBS study (J. P.) was supported by the Finnish Diabetes Research Foundation, Kuopio University Hospital Project grant (EVO/VTR grants 2005-2019), and the Academy of Finland grant (Contract no. 138006). This study was funded by the European Research Council under the European Union's Horizon 2020 research and innovation program (Grant No. 802825 to M. U. K.). K. H. P. was funded by the Academy of Finland (grant numbers 272376, 266286, 314383, and 335443), the Finnish Medical Foundation, Gyllenberg Foundation, Novo Nordisk Foundation (grant numbers NNF10OC1013354, NNF17OC0027232, and NNF20OC0060547), Finnish Diabetes Research Foundation, Finnish Foundation for Cardiovascular Research, University of Helsinki, and Helsinki University Hospital and Government Research Funds. I. S. was funded by the Instrumentarium Science Foundation. Personal grants to U. T. A. were received from the Matti and Vappu Maukonen Foundation, Ella och Georg Ehrnrooths Stiftelse and the Finnish Foundation for Cardiovascular Research.
Keywords: Adipogenesis; Dual-tissue transcriptomics screening; Liver histology; Non-alcoholic fatty liver disease; Obesity; Serum biomarkers; cis regulatory variants.
Copyright © 2023 The Author(s). Published by Elsevier B.V. All rights reserved.
Conflict of interest statement
Declaration of interests J. N. B. received consulting fees from GLG, and support for attending meetings and/or travel from the American Gastroenterology Association (AGA), at least once during the last 36 months. The other authors declare that they have no competing interests.
Figures


References
-
- Younossi Z.M., Koenig A.B., Abdelatif D., Fazel Y., Henry L., Wymer M. Global epidemiology of nonalcoholic fatty liver disease—meta-analytic assessment of prevalence, incidence, and outcomes. Hepatology. 2016;64(1):73–84. - PubMed
-
- Powell E.E., Wong V.W.S., Rinella M. Non-alcoholic fatty liver disease. Lancet. 2021;397(10290):2212–2224. - PubMed
-
- Wong R.J., Aguilar M., Cheung R., et al. Nonalcoholic steatohepatitis is the second leading etiology of liver disease among adults awaiting liver transplantation in the United States. Gastroenterology. 2015;148(3):547–555. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

