Comparative analysis of Tet2 catalytic-deficient and knockout bone marrow over time
- PMID: 37225048
- PMCID: PMC10524687
- DOI: 10.1016/j.exphem.2023.05.004
Comparative analysis of Tet2 catalytic-deficient and knockout bone marrow over time
Abstract
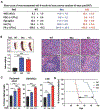
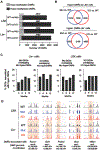
TET2 is a member of the Ten-eleven translocation (Tet) family of DNA dioxygenases that regulate gene expression by promoting DNA demethylation (enzymatic activity) and partnering with chromatin regulatory complexes (nonenzymatic functions). TET2 is highly expressed in the hematopoietic lineage, where its molecular functions are the subject of continuous investigations because of the prevalence of TET2 mutations in hematologic malignancies. Previously, we have implicated Tet2 catalytic and noncatalytic functions in the regulation of myeloid and lymphoid lineages, respectively. However, the impact of these functions of Tet2 on hematopoiesis as the bone marrow ages remains unclear. Here, we conducted comparative transplantations and transcriptomic analyses of 3-, 6-, 9-, and 12-month-old Tet2 catalytic mutant (Mut) and knockout (KO) bone marrow. Tet2 Mut bone marrow of all ages exclusively caused hematopoietic disorders of the myeloid lineage. In contrast, young Tet2 KO bone marrow developed both lymphoid and myeloid diseases, whereas older Tet2 KO bone marrow predominantly elicited myeloid disorders with shorter latency than age-matched Tet2 Mut bone marrow. We identified robust gene dysregulation in Tet2 KO Lin- cells at 6 months that involved lymphoma and myelodysplastic syndrome and/or leukemia-causing genes, many of which were hypermethylated early in life. There was a shift from lymphoid to myeloid gene deregulation in Tet2 KO Lin- cells with age, underpinning the higher incidence of myeloid diseases. These findings expand on the dynamic regulation of bone marrow by Tet2 and show that its catalytic-dependent and -independent roles have distinct impacts on myeloid and lymphoid lineages with age.
Copyright © 2023 ISEH -- Society for Hematology and Stem Cells. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Disclosure of conflict of interests
The authors declare no competing financial or other conflicts of interest.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

