Gut microbiome signatures of extreme environment adaption in Tibetan pig
- PMID: 37225687
- PMCID: PMC10209067
- DOI: 10.1038/s41522-023-00395-3
Gut microbiome signatures of extreme environment adaption in Tibetan pig
Abstract
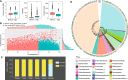
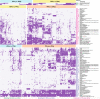
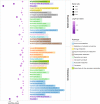
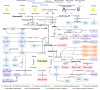
Tibetan pigs (TPs) can adapt to the extreme environments in the Tibetan plateau implicated by their self-genome signals, but little is known about roles of the gut microbiota in the host adaption. Here, we reconstructed 8210 metagenome-assembled genomes from TPs (n = 65) living in high-altitude and low-altitude captive pigs (87 from China-CPs and 200 from Europe-EPs) that were clustered into 1050 species-level genome bins (SGBs) at the threshold of 95% average nucleotide identity. 73.47% of SGBs represented new species. The gut microbial community structure analysis based on 1,048 SGBs showed that TPs was significantly different from low-altitude captive pigs. TP-associated SGBs enabled to digest multiple complex polysaccharides, including cellulose, hemicellulose, chitin and pectin. Especially, we found TPs showed the most common enrichment of phyla Fibrobacterota and Elusimicrobia, which were involved in the productions of short- and medium-chain fatty acids (acetic acid, butanoate and propanoate; octanomic, decanoic and dodecanoic acids), as well as in the biosynthesis of lactate, 20 essential amino acids, multiple B vitamins (B1, B2, B3, B5, B7 and B9) and cofactors. Unexpectedly, Fibrobacterota solely showed powerful metabolic capacity, including the synthesis of acetic acid, alanine, histidine, arginine, tryptophan, serine, threonine, valine, B2, B5, B9, heme and tetrahydrofolate. These metabolites might contribute to host adaptation to high-altitude, such as energy harvesting and resistance against hypoxia and ultraviolet radiation. This study provides insights into understanding the role of gut microbiome played in mammalian high-altitude adaptation and discovers some potential microbes as probiotics for improving animal health.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures

References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

