A ZFYVE21-Rubicon-RNF34 signaling complex promotes endosome-associated inflammasome activity in endothelial cells
- PMID: 37225719
- PMCID: PMC10209169
- DOI: 10.1038/s41467-023-38684-2
A ZFYVE21-Rubicon-RNF34 signaling complex promotes endosome-associated inflammasome activity in endothelial cells
Erratum in
-
Author Correction: A ZFYVE21-Rubicon-RNF34 signaling complex promotes endosome-associated inflammasome activity in endothelial cells.Nat Commun. 2023 Jun 7;14(1):3336. doi: 10.1038/s41467-023-39154-5. Nat Commun. 2023. PMID: 37286577 Free PMC article. No abstract available.
-
Author Correction: A ZFYVE21-Rubicon-RNF34 signaling complex promotes endosome-associated inflammasome activity in endothelial cells.Nat Commun. 2023 Dec 18;14(1):8404. doi: 10.1038/s41467-023-44225-8. Nat Commun. 2023. PMID: 38110370 Free PMC article. No abstract available.
Abstract
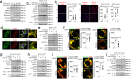
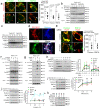
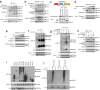
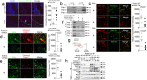
Internalization of complement membrane attack complexes (MACs) assembles NLRP3 inflammasomes in endothelial cells (EC) and promotes IL-β-mediated tissue inflammation. Informed by proteomics analyses of FACS-sorted inflammasomes, we identify a protein complex modulating inflammasome activity on endosomes. ZFVYE21, a Rab5 effector, partners with Rubicon and RNF34, forming a "ZRR" complex that is stabilized in a Rab5- and ZFYVE21-dependent manner on early endosomes. There, Rubicon competitively disrupts inhibitory associations between caspase-1 and its pseudosubstrate, Flightless I (FliI), while RNF34 ubiquitinylates and degradatively removes FliI from the signaling endosome. The concerted actions of the ZRR complex increase pools of endosome-associated caspase-1 available for activation. The ZRR complex is assembled in human tissues, its associated signaling responses occur in three mouse models in vivo, and the ZRR complex promotes inflammation in a skin model of chronic rejection. The ZRR signaling complex reflects a potential therapeutic target for attenuating inflammasome-mediated tissue injury.
© 2023. This is a U.S. Government work and not under copyright protection in the US; foreign copyright protection may apply.
Conflict of interest statement
The authors declare no competing interests.
Figures




References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

