Heteromeric clusters of ubiquitinated ER-shaping proteins drive ER-phagy
- PMID: 37225994
- PMCID: PMC10247384
- DOI: 10.1038/s41586-023-06090-9
Heteromeric clusters of ubiquitinated ER-shaping proteins drive ER-phagy
Abstract
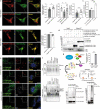
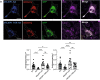
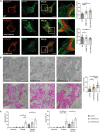
Membrane-shaping proteins characterized by reticulon homology domains play an important part in the dynamic remodelling of the endoplasmic reticulum (ER). An example of such a protein is FAM134B, which can bind LC3 proteins and mediate the degradation of ER sheets through selective autophagy (ER-phagy)1. Mutations in FAM134B result in a neurodegenerative disorder in humans that mainly affects sensory and autonomic neurons2. Here we report that ARL6IP1, another ER-shaping protein that contains a reticulon homology domain and is associated with sensory loss3, interacts with FAM134B and participates in the formation of heteromeric multi-protein clusters required for ER-phagy. Moreover, ubiquitination of ARL6IP1 promotes this process. Accordingly, disruption of Arl6ip1 in mice causes an expansion of ER sheets in sensory neurons that degenerate over time. Primary cells obtained from Arl6ip1-deficient mice or from patients display incomplete budding of ER membranes and severe impairment of ER-phagy flux. Therefore, we propose that the clustering of ubiquitinated ER-shaping proteins facilitates the dynamic remodelling of the ER during ER-phagy and is important for neuronal maintenance.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures












References
-
- Khaminets A, et al. Regulation of endoplasmic reticulum turnover by selective autophagy. Nature. 2015;522:354–358. - PubMed
-
- Kurth I, et al. Mutations in FAM134B, encoding a newly identified Golgi protein, cause severe sensory and autonomic neuropathy. Nat. Genet. 2009;41:1179–1181. - PubMed
-
- Mochida K, et al. Receptor-mediated selective autophagy degrades the endoplasmic reticulum and the nucleus. Nature. 2015;522:359–362. - PubMed
-
- Hübner CA, Kurth I. Membrane-shaping disorders: a common pathway in axon degeneration. Brain. 2014;137:3109–3121. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

