Microbial carbon use efficiency promotes global soil carbon storage
- PMID: 37225998
- PMCID: PMC10307633
- DOI: 10.1038/s41586-023-06042-3
Microbial carbon use efficiency promotes global soil carbon storage
Abstract
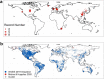
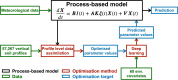
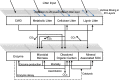
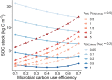
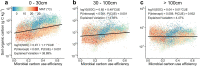
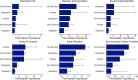
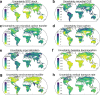
Soils store more carbon than other terrestrial ecosystems1,2. How soil organic carbon (SOC) forms and persists remains uncertain1,3, which makes it challenging to understand how it will respond to climatic change3,4. It has been suggested that soil microorganisms play an important role in SOC formation, preservation and loss5-7. Although microorganisms affect the accumulation and loss of soil organic matter through many pathways4,6,8-11, microbial carbon use efficiency (CUE) is an integrative metric that can capture the balance of these processes12,13. Although CUE has the potential to act as a predictor of variation in SOC storage, the role of CUE in SOC persistence remains unresolved7,14,15. Here we examine the relationship between CUE and the preservation of SOC, and interactions with climate, vegetation and edaphic properties, using a combination of global-scale datasets, a microbial-process explicit model, data assimilation, deep learning and meta-analysis. We find that CUE is at least four times as important as other evaluated factors, such as carbon input, decomposition or vertical transport, in determining SOC storage and its spatial variation across the globe. In addition, CUE shows a positive correlation with SOC content. Our findings point to microbial CUE as a major determinant of global SOC storage. Understanding the microbial processes underlying CUE and their environmental dependence may help the prediction of SOC feedback to a changing climate.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures
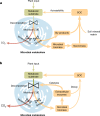
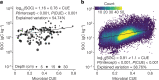
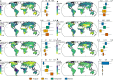
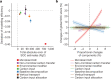
Comment in
-
Model uncertainty obscures major driver of soil carbon.Nature. 2024 Mar;627(8002):E1-E3. doi: 10.1038/s41586-023-06999-1. Epub 2024 Mar 6. Nature. 2024. PMID: 38448702 No abstract available.
References
-
- Jackson RB, et al. The ecology of soil carbon: pools, vulnerabilities, and biotic and abiotic controls. Annu. Rev. Ecol. Evol. Syst. 2017;48:419–445. doi: 10.1146/annurev-ecolsys-112414-054234. - DOI
-
- Ciais, P. et al. in IPCC Climate Change 2013: The Physical Science Basis (eds Stocker, T. F. et al.) 465–570 (Cambridge Univ. Press, 2014).
-
- Bradford MA, et al. Managing uncertainty in soil carbon feedbacks to climate change. Nat. Clim. Change. 2016;6:751–758. doi: 10.1038/nclimate3071. - DOI
-
- Cotrufo MF, Wallenstein MD, Boot CM, Denef K, Paul E. The microbial efficiency‐matrix stabilization (MEMS) framework integrates plant litter decomposition with soil organic matter stabilization: do labile plant inputs form stable soil organic matter? Glob. Change Biol. 2013;19:988–995. doi: 10.1111/gcb.12113. - DOI - PubMed

