Employing computational tools to design a multi-epitope vaccine targeting human immunodeficiency virus-1 (HIV-1)
- PMID: 37226084
- PMCID: PMC10206567
- DOI: 10.1186/s12864-023-09330-4
Employing computational tools to design a multi-epitope vaccine targeting human immunodeficiency virus-1 (HIV-1)
Abstract
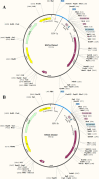
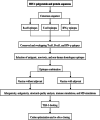
Background: Despite being in the 21st century, the world has still not been able to vanquish the global AIDS epidemic, and the only foreseeable solution seems to be a safe and effective vaccine. Unfortunately, vaccine trials so far have returned unfruitful results, possibly due to their inability to induce effective cellular, humoral and innate immune responses. The current study aims to tackle these limitations and propose the desired vaccine utilizing immunoinformatic approaches that have returned promising results in designing vaccines against various rapidly mutating organisms. For this, all polyprotein and protein sequences of HIV-1 were retrieved from the LANL (Los Alamos National Laboratory) database. The consensus sequence was generated after alignment and used to predict epitopes. Conserved, antigenic, non-allergenic, T-cell inducing, B-cell inducing, IFN-ɣ inducing, non-human homologous epitopes were selected and combined to propose two vaccine constructs i.e., HIV-1a (without adjuvant) and HIV-1b (with adjuvant).
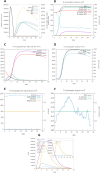
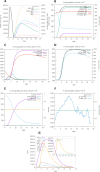
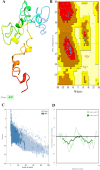
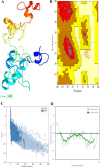
Results: HIV-1a and HIV-1b were subjected to antigenicity, allergenicity, structural quality analysis, immune simulations, and MD (molecular dynamics) simulations. Both proposed multi-epitope vaccines were found to be antigenic, non-allergenic, stable, and induce cellular, humoral, and innate immune responses. TLR-3 docking and in-silico cloning of both constructs were also performed.
Conclusion: Our results indicate HIV-1b to be more promising than HIV-1a; experimental validations can confirm the efficacy and safety of both constructs and in-vivo efficacy in animal models.
Keywords: Acquired immunodeficiency syndrome; Bioinformatics; Computational biology; Human immunodeficiency virus; Immunity; Immuno-informatics; Toll like receptor-3; Vaccinology.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures

Similar articles
-
Exploring T & B-cell epitopes and designing multi-epitope subunit vaccine targeting integration step of HIV-1 lifecycle using immunoinformatics approach.Microb Pathog. 2019 Dec;137:103791. doi: 10.1016/j.micpath.2019.103791. Epub 2019 Oct 10. Microb Pathog. 2019. PMID: 31606417
-
Immunoinformatic-driven design and evaluation of multi-epitope mRNA vaccine targeting HIV-1 gp120.Front Immunol. 2025 May 13;16:1480025. doi: 10.3389/fimmu.2025.1480025. eCollection 2025. Front Immunol. 2025. PMID: 40433366 Free PMC article.
-
Reverse vaccinology approaches to design a potent multiepitope vaccine against the HIV whole genome: immunoinformatic, bioinformatics, and molecular dynamics approaches.BMC Infect Dis. 2024 Aug 28;24(1):873. doi: 10.1186/s12879-024-09775-2. BMC Infect Dis. 2024. PMID: 39198721 Free PMC article.
-
Multiple Approaches for Increasing the Immunogenicity of an Epitope-Based Anti-HIV Vaccine.AIDS Res Hum Retroviruses. 2015 Nov;31(11):1077-88. doi: 10.1089/AID.2015.0101. Epub 2015 Aug 13. AIDS Res Hum Retroviruses. 2015. PMID: 26149745 Review.
-
Better Epitope Discovery, Precision Immune Engineering, and Accelerated Vaccine Design Using Immunoinformatics Tools.Front Immunol. 2020 Apr 7;11:442. doi: 10.3389/fimmu.2020.00442. eCollection 2020. Front Immunol. 2020. PMID: 32318055 Free PMC article. Review.
Cited by
-
Designing a Multi-Epitope Vaccine Against MPXV and HIV Based on an Immunoinformatic Approach.Int J Mol Sci. 2025 Jun 30;26(13):6313. doi: 10.3390/ijms26136313. Int J Mol Sci. 2025. PMID: 40650090 Free PMC article.
-
Immunoinformatic Identification of Multiple Epitopes of gp120 Protein of HIV-1 to Enhance the Immune Response against HIV-1 Infection.Int J Mol Sci. 2024 Feb 19;25(4):2432. doi: 10.3390/ijms25042432. Int J Mol Sci. 2024. PMID: 38397105 Free PMC article.
-
Design of multivalent-epitope vaccine models directed toward the world's population against HIV-Gag polyprotein: Reverse vaccinology and immunoinformatics.PLoS One. 2024 Sep 27;19(9):e0306559. doi: 10.1371/journal.pone.0306559. eCollection 2024. PLoS One. 2024. PMID: 39331650 Free PMC article.
-
Integrated Immuno and bioinformatics assisted novel epitope vaccine against HIV infection: a study based on complete genome.Virol J. 2025 Jul 9;22(1):228. doi: 10.1186/s12985-025-02764-0. Virol J. 2025. PMID: 40634976 Free PMC article.
-
Revisiting the potential of natural antimicrobial peptides against emerging respiratory viral disease: a review.3 Biotech. 2025 Feb;15(2):40. doi: 10.1007/s13205-024-04184-3. Epub 2025 Jan 13. 3 Biotech. 2025. PMID: 39816617 Review.
References
-
- Data U. Available online: https://www.unaids.org/sites/default/files/media_asset2020.
-
- Organization WH. Policy brief: update of recommendations on first-and second-line antiretroviral regimens. Geneva: World Health Organization; 2019.
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

