Drug mechanism enrichment analysis improves prioritization of therapeutics for repurposing
- PMID: 37226094
- PMCID: PMC10207828
- DOI: 10.1186/s12859-023-05343-8
Drug mechanism enrichment analysis improves prioritization of therapeutics for repurposing
Abstract
Background: There is a pressing need for improved methods to identify effective therapeutics for diseases. Many computational approaches have been developed to repurpose existing drugs to meet this need. However, these tools often output long lists of candidate drugs that are difficult to interpret, and individual drug candidates may suffer from unknown off-target effects. We reasoned that an approach which aggregates information from multiple drugs that share a common mechanism of action (MOA) would increase on-target signal compared to evaluating drugs on an individual basis. In this study, we present drug mechanism enrichment analysis (DMEA), an adaptation of gene set enrichment analysis (GSEA), which groups drugs with shared MOAs to improve the prioritization of drug repurposing candidates.
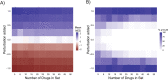
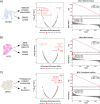
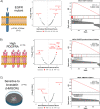
Results: First, we tested DMEA on simulated data and showed that it can sensitively and robustly identify an enriched drug MOA. Next, we used DMEA on three types of rank-ordered drug lists: (1) perturbagen signatures based on gene expression data, (2) drug sensitivity scores based on high-throughput cancer cell line screening, and (3) molecular classification scores of intrinsic and acquired drug resistance. In each case, DMEA detected the expected MOA as well as other relevant MOAs. Furthermore, the rankings of MOAs generated by DMEA were better than the original single-drug rankings in all tested data sets. Finally, in a drug discovery experiment, we identified potential senescence-inducing and senolytic drug MOAs for primary human mammary epithelial cells and then experimentally validated the senolytic effects of EGFR inhibitors.
Conclusions: DMEA is a versatile bioinformatic tool that can improve the prioritization of candidates for drug repurposing. By grouping drugs with a shared MOA, DMEA increases on-target signal and reduces off-target effects compared to analysis of individual drugs. DMEA is publicly available as both a web application and an R package at https://belindabgarana.github.io/DMEA .
Keywords: Drug repurposing; Enrichment analysis; Gene expression analysis; Mechanism of action; Precision medicine; Proteomic analysis; Senescence; Senolytic; Targeted therapeutics.
© 2023. The Author(s).
Conflict of interest statement
The authors declare that they have no competing interests.
Figures



Similar articles
-
Open MoA: revealing the mechanism of action (MoA) based on network topology and hierarchy.Bioinformatics. 2023 Nov 1;39(11):btad666. doi: 10.1093/bioinformatics/btad666. Bioinformatics. 2023. PMID: 37930015 Free PMC article.
-
Deep learning large-scale drug discovery and repurposing.Nat Comput Sci. 2024 Aug;4(8):600-614. doi: 10.1038/s43588-024-00679-4. Epub 2024 Aug 21. Nat Comput Sci. 2024. PMID: 39169261
-
MD-Miner: a network-based approach for personalized drug repositioning.BMC Syst Biol. 2017 Oct 3;11(Suppl 5):86. doi: 10.1186/s12918-017-0462-9. BMC Syst Biol. 2017. PMID: 28984195 Free PMC article.
-
Computational Drug Repurposing: Current Trends.Curr Med Chem. 2019;26(28):5389-5409. doi: 10.2174/0929867325666180530100332. Curr Med Chem. 2019. PMID: 29848268 Review.
-
Repurposing digoxin for geroprotection in patients with frailty and multimorbidity.Ageing Res Rev. 2023 Apr;86:101860. doi: 10.1016/j.arr.2023.101860. Epub 2023 Jan 20. Ageing Res Rev. 2023. PMID: 36682465 Review.
Cited by
-
Reconciling multiple connectivity-based systems biology methods for drug repurposing.Brief Bioinform. 2025 Jul 2;26(4):bbaf387. doi: 10.1093/bib/bbaf387. Brief Bioinform. 2025. PMID: 40736744 Free PMC article. Review.
-
PARP Inhibition in Colorectal Cancer-A Comparison of Potential Predictive Biomarkers for Therapy.Pharmaceuticals (Basel). 2025 Jun 17;18(6):905. doi: 10.3390/ph18060905. Pharmaceuticals (Basel). 2025. PMID: 40573300 Free PMC article.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

