Recent advances in mycobacterial membrane protein large 3 inhibitor drug design for mycobacterial infections
- PMID: 37226498
- PMCID: PMC10330604
- DOI: 10.1080/17460441.2023.2218082
Recent advances in mycobacterial membrane protein large 3 inhibitor drug design for mycobacterial infections
Abstract
Introduction: Tuberculosis and nontuberculous mycobacterial infections are notoriously difficult to treat, requiring long-courses of intensive multi-drug therapies associated with adverse side effects. To identify better therapeutics, whole cell screens have identified novel pharmacophores, a surprisingly high number of which target an essential lipid transporter known as MmpL3.
Areas covered: This paper summarizes what is known about MmpL3, its mechanism of lipid transport and therapeutic potential, and provides an overview of the different classes of MmpL3 inhibitors currently under development. It further describes the assays available to study MmpL3 inhibition by these compounds.
Expert opinion: MmpL3 has emerged as a target of high therapeutic value. Accordingly, several classes of MmpL3 inhibitors are currently under development with one drug candidate (SQ109) having undergone a Phase 2b clinical study. The hydrophobic character of most MmpL3 series identified to date seems to drive antimycobacterial potency resulting in poor bioavailability, which is a significant impediment to their development. There is also a need for more high-throughput and informative assays to elucidate the precise mechanism of action of MmpL3 inhibitors and drive the rational optimization of analogues.
Keywords: MmpL3; Mycobacterium; drug development; mycolic acids; non-tuberculous mycobacteria; tuberculosis.
Conflict of interest statement
Declaration of Interest
The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
Figures

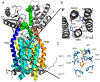

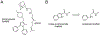
References
-
- World Health Organization: Global Tuberculosis Report 2021. 2021.
-
- Fernandes GFS, Thompson AM, Castagnolo D, Denny WA, Dos Santos JL: Tuberculosis Drug Discovery: Challenges and New Horizons. J Med Chem 2022, 65:7489–7531. - PubMed
-
- Lee BS, Pethe K: Therapeutic potential of promiscuous targets in Mycobacterium tuberculosis. Curr Opin Pharmacol 2018, 42:22–26. - PubMed
-
- Goldman RC: Why are membrane targets discovered by phenotypic screens and genome sequencing in Mycobacterium tuberculosis? Tuberculosis (Edinb) 2013, 93:569–588. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
