Folding of Staphylococcal Nuclease Induced by Binding of Chemically Modified Substrate Analogues Sheds Light on Mechanisms of Coupled Folding/Binding Reactions
- PMID: 37227385
- PMCID: PMC10583223
- DOI: 10.1021/acs.biochem.3c00094
Folding of Staphylococcal Nuclease Induced by Binding of Chemically Modified Substrate Analogues Sheds Light on Mechanisms of Coupled Folding/Binding Reactions
Abstract
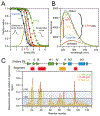
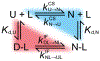
Several proteins have been shown to undergo a shift in the mechanism of ligand binding-induced folding from conformational selection (CS; folding precedes binding) to induced fit (IF; binding precedes folding) with increasing ligand concentration. In previous studies of the coupled folding/binding reaction of staphylococcal nuclease (SNase) in the presence of a substrate analogue, adenosine-3',5'-diphosphate (prAp), we found that the two phosphate groups make important energetic contributions toward stabilizing its complex with the native protein as well as transient conformational states encountered at high ligand concentrations favoring IF. However, the structural contributions of each phosphate group during the reaction remain unclear. To address this question, we relied on fluorescence, nuclear magnetic resonance (NMR), absorption, and isothermal titration calorimetry to study the effects of deletion of the phosphate groups of prAp on the kinetics of ligand-induced folding, using a strategy analogous to mutational ϕ-value analysis to interpret the results. Kinetic measurements over a wide range of ligand concentrations, together with structural characterization of a transient protein-ligand encounter complex using 2D NMR, indicated that, at high ligand concentrations favoring IF, (i) the 5'-phosphate group interacts weakly with denatured SNase during early stages of the reaction, resulting in loose docking of the two domains of SNase, and (ii) the 3'-phosphate group engages in some specific contacts with the polypeptide in the transition state prior to formation of the native SNase-prAp complex.
Figures
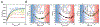


Similar articles
-
Direct comparison of the structural dynamics between spontaneous and ligand-induced folding of staphylococcal nuclease.Protein Sci. 2025 May;34(5):e70135. doi: 10.1002/pro.70135. Protein Sci. 2025. PMID: 40285686
-
Energetics and kinetics of substrate analog-coupled staphylococcal nuclease folding revealed by a statistical mechanical approach.Proc Natl Acad Sci U S A. 2020 Aug 18;117(33):19953-19962. doi: 10.1073/pnas.1914349117. Epub 2020 Jul 31. Proc Natl Acad Sci U S A. 2020. PMID: 32737158 Free PMC article.
-
Effect of salts on the stability and folding of staphylococcal nuclease.Biochemistry. 2001 Feb 20;40(7):2113-28. doi: 10.1021/bi000861k. Biochemistry. 2001. PMID: 11329280
-
Thermodynamics of denaturation of staphylococcal nuclease mutants: an intermediate state in protein folding.FASEB J. 1996 Jan;10(1):67-74. doi: 10.1096/fasebj.10.1.8566550. FASEB J. 1996. PMID: 8566550 Review.
-
Staphylococcal nuclease reviewed: a prototypic study in contemporary enzymology. IV. The nuclease as a model for protein folding.Mol Cell Biochem. 1979 Feb 9;23(3):131-41. doi: 10.1007/BF00219452. Mol Cell Biochem. 1979. PMID: 90333 Review.
Cited by
-
Direct comparison of the structural dynamics between spontaneous and ligand-induced folding of staphylococcal nuclease.Protein Sci. 2025 May;34(5):e70135. doi: 10.1002/pro.70135. Protein Sci. 2025. PMID: 40285686
-
A Kinetic Transition Network Model Reveals the Diversity of Protein Dimer Formation Mechanisms.Biomolecules. 2023 Nov 26;13(12):1708. doi: 10.3390/biom13121708. Biomolecules. 2023. PMID: 38136580 Free PMC article.
-
Protein structure-function continuum model: Emerging nexuses between specificity, evolution, and structure.Protein Sci. 2024 Apr;33(4):e4968. doi: 10.1002/pro.4968. Protein Sci. 2024. PMID: 38532700 Free PMC article. Review.
References
-
- Perutz MF (1970) Stereochemistry of cooperative effects in haemoglobin, Nature 228, 726739. - PubMed
-
- Müller CW, Schlauderer GJ, Reinstein J, and Schulz GE (1996) Adenylate kinase motions during catalysis: an energetic counterweight balancing substrate binding, Structure 4, 147–156. - PubMed
-
- Ferguson KM, Berger MB, Mendrola JM, Cho HS, Leahy DJ, and Lemmon MA (2003) EGF activates its receptor by removing interactions that autoinhibit ectodomain dimerization, Mol. Cell 11, 507–517. - PubMed
-
- Wright PE, and Dyson HJ (1999) Intrinsically unstructured proteins: re-assessing the protein structure-function paradigm, J. Mol. Biol 293, 321–331. - PubMed
-
- Dunker AK, Lawson JD, Brown CJ, Williams RM, Romero P, Oh JS, Oldfield CJ, Campen AM, Ratliff CM, Hipps KW, Ausio J, Nissen MS, Reeves R, Kang C, Kissinger CR, Bailey RW, Griswold MD, Chiu W, Garner EC, and Obradovic Z (2001) Intrinsically disordered protein, J. Mol. Graph. Model 19, 26–59. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

