Cholesterol Contributes to Risk, Severity, and Machine Learning-Driven Diagnosis of Lyme Disease
- PMID: 37227948
- PMCID: PMC10506776
- DOI: 10.1093/cid/ciad307
Cholesterol Contributes to Risk, Severity, and Machine Learning-Driven Diagnosis of Lyme Disease
Abstract
Background: Lyme disease is the most prevalent vector-borne disease in the US, yet its host factors are poorly understood and diagnostic tests are limited. We evaluated patients in a large health system to uncover cholesterol's role in the susceptibility, severity, and machine learning-based diagnosis of Lyme disease.
Methods: A longitudinal health system cohort comprised 1 019 175 individuals with electronic health record data and 50 329 with linked genetic data. Associations of blood cholesterol level, cholesterol genetic scores comprising common genetic variants, and burden of rare loss-of-function (LoF) variants in cholesterol metabolism genes with Lyme disease were investigated. A portable machine learning model was constructed and tested to predict Lyme disease using routine lipid and clinical measurements.
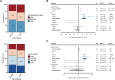
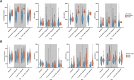
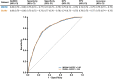
Results: There were 3832 cases of Lyme disease. Increasing cholesterol was associated with greater risk of Lyme disease and hypercholesterolemia was more prevalent in Lyme disease cases than in controls. Cholesterol genetic scores and rare LoF variants in CD36 and LDLR were associated with Lyme disease risk. Serological profiling of cases revealed parallel trajectories of rising cholesterol and immunoglobulin levels over the disease course, including marked increases in individuals with LoF variants and high cholesterol genetic scores. The machine learning model predicted Lyme disease solely using routine lipid panel, blood count, and metabolic measurements.
Conclusions: These results demonstrate the value of large-scale genetic and clinical data to reveal host factors underlying infectious disease biology, risk, and prognosis and the potential for their clinical translation to machine learning diagnostics that do not need specialized assays.
Keywords: Lyme disease; cholesterol; exome sequencing; machine learning.
© The Author(s) 2023. Published by Oxford University Press on behalf of Infectious Diseases Society of America. All rights reserved. For permissions, please e-mail: journals.permissions@oup.com.
Conflict of interest statement
Potential conflicts of interest. R. D. reports grants from AstraZeneca; grants and nonfinancial support from Goldfinch Bio; being a scientific cofounder, consultant, and equity holder for Pensieve Health; and being a consultant for Variant Bio, all not related to this work. All remaining authors: No reported conflicts of interest. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
Figures

References
-
- Mead PS. Epidemiology of Lyme disease. Infect Dis Clin North Am 2015; 29:187–210. - PubMed
-
- Nowakowski J, McKenna D, Nadelman RB, et al. . Failure of treatment with cephalexin for Lyme disease. Arch Fam Med 2000; 9:563–7. - PubMed
-
- Fallon BA, Kochevar JM, Gaito A, Nields JA. The underdiagnosis of neuropsychiatric Lyme disease in children and adults. Psychiatr Clin North Am 1998; 21:693–703. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

