Substance P analogs devoid of key residues fail to activate human mast cells via MRGPRX2
- PMID: 37228611
- PMCID: PMC10203606
- DOI: 10.3389/fimmu.2023.1155740
Substance P analogs devoid of key residues fail to activate human mast cells via MRGPRX2
Abstract
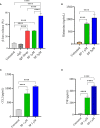
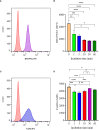
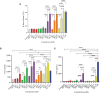
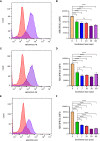
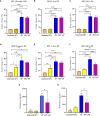
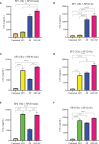
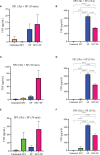
Mast cells play an important role in disease pathogenesis by secreting immunomodulatory molecules. Mast cells are primarily activated by the crosslinking of their high affinity IgE receptors (FcεRI) by antigen bound immunoglobulin (Ig)E antibody complexes. However, mast cells can also be activated by the mas related G protein-coupled receptor X2 (MRGPRX2), in response to a range of cationic secretagogues, such as substance P (SP), which is associated with pseudo-allergic reactions. We have previously reported that the in vitro activation of mouse mast cells by basic secretagogues is mediated by the mouse orthologue of the human MRGPRX2, MRGPRB2. To further elucidate the mechanism of MRGPRX2 activation, we studied the time-dependent internalization of MRGPRX2 by human mast cells (LAD2) upon stimulation with the neuropeptide SP. In addition, we performed computational studies to identify the intermolecular forces that facilitate ligand-MRGPRX2 interaction using SP. The computational predictions were tested experimentally by activating LAD2 with SP analogs, which were missing key amino acid residues. Our data suggest that mast cell activation by SP causes internalization of MRGPRX2 within 1 min of stimulation. Hydrogen bonds (h-bonds) and salt bridges govern the biding of SP to MRGPRX2. Arg1 and Lys3 in SP are key residues that are involved in both h-bonding and salt bridge formations with Glu164 and Asp184 of MRGPRX2, respectively. In accordance, SP analogs devoid of key residues (SP1 and SP2) failed to activate MRGPRX2 degranulation. However, both SP1 and SP2 caused a comparable release of chemokine CCL2. Further, SP analogs SP1, SP2 and SP4 did not activate tumor necrosis factor (TNF) production. We further show that SP1 and SP2 limit the activity of SP on mast cells. The results provide important mechanistic insight into the events that result in mast cell activation through MRGPRX2 and highlight the important physiochemical characteristics of a peptide ligand that facilitates ligand-MRGPRX2 interactions. The results are important in understanding activation through MRGPRX2, and the intermolecular forces that govern ligand-MRGPRX2 interaction. The elucidation of important physiochemical properties within a ligand that are needed for receptor interaction will aid in designing novel therapeutics and antagonists for MRGPRX2.
Keywords: MRGPRX2; amino acid residues; ligand-receptor interactions; mast cells; substance P.
Copyright © 2023 Raj, Hlushak, Arizmendi, Kovalenko and Kulka.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

Similar articles
-
Inhibition of MRGPRX2 but not FcεRI or MrgprB2-mediated mast cell degranulation by a small molecule inverse receptor agonist.Front Immunol. 2022 Oct 6;13:1033794. doi: 10.3389/fimmu.2022.1033794. eCollection 2022. Front Immunol. 2022. PMID: 36275683 Free PMC article.
-
Caffeic acid phenethyl ester inhibits pseudo-allergic reactions via inhibition of MRGPRX2/MrgprB2-dependent mast cell degranulation.Arch Pharm Res. 2022 Sep;45(9):644-657. doi: 10.1007/s12272-022-01405-2. Epub 2022 Oct 2. Arch Pharm Res. 2022. PMID: 36183260
-
Lactic acid suppresses MRGPRX2 mediated mast cell responses.Cell Immunol. 2021 Oct;368:104422. doi: 10.1016/j.cellimm.2021.104422. Epub 2021 Aug 8. Cell Immunol. 2021. PMID: 34399172 Free PMC article.
-
Unlocking the Non-IgE-Mediated Pseudo-Allergic Reaction Puzzle with Mas-Related G-Protein Coupled Receptor Member X2 (MRGPRX2).Cells. 2021 Apr 27;10(5):1033. doi: 10.3390/cells10051033. Cells. 2021. PMID: 33925682 Free PMC article. Review.
-
MRGPRX2 sensing of cationic compounds-A bridge between nociception and skin diseases?Exp Dermatol. 2021 Feb;30(2):193-200. doi: 10.1111/exd.14222. Epub 2020 Nov 17. Exp Dermatol. 2021. PMID: 33107136 Review.
Cited by
-
Itch: from the skin to the brain - peripheral and central neural sensitization in chronic itch.Front Mol Neurosci. 2023 Oct 2;16:1272230. doi: 10.3389/fnmol.2023.1272230. eCollection 2023. Front Mol Neurosci. 2023. PMID: 37849619 Free PMC article. Review.
-
Cellular and Molecular Mechanisms of Mast Cells in Atherosclerotic Plaque Progression and Destabilization.Clin Rev Allergy Immunol. 2024 Feb;66(1):30-49. doi: 10.1007/s12016-024-08981-9. Epub 2024 Jan 30. Clin Rev Allergy Immunol. 2024. PMID: 38289515 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

